Abstract
Most biological discoveries can only be made in light of evolution. In particular, functional annotation of genes is usually deduced from the orthology, paralogy, or xenology relations between genes, which are inferred from the comparison of a gene tree with a species tree. As sequence-only gene tree reconstruction methods often do not allow to confidently discriminate between trees, recent “integrative methods” include information from the species tree. The idea is to consider, in addition to a value measuring the fitness of a tree to a sequence alignment, a measure reflecting the evolution of a whole gene family through gene gain and loss. One such measure is the “reconciliation” cost, i.e., the cost of a gain and loss scenario explaining the incongruence between the gene and species tree. This chapter begins with a review of deterministic algorithms for computing reconciliation distances under various evolutionary models of gene family evolution. We then review integrative methods for correcting a gene tree, based on various strategies for exploring its neighborhood. The considered algorithms are those based on polytomy resolution, tree amalgamation and supertree reconstruction. The goal is to provide a comprehensive overview of existing methods with algorithms presented in concise form. The reader is referred to original papers for more details and proofs of complexity.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
5.1 Introduction
Genes are the molecular units of heredity holding the information to build and maintain cells. They are key to understanding biological mechanisms, identifying genetic variation, and designing appropriate gene therapies.
In the course of evolution, genes are mutated, duplicated, lost, and passed to organisms through speciation or Horizontal Gene Transfer (HGT), the exchange of genetic material among coexisting species. Therefore, most biological discoveries can only be made in the light of evolution. Genes originating from the same ancestral copy are called homologs. Homologous genes are grouped into gene families, usually via sequence similarity methods. Moreover, they can be orthologs if their most recent common ancestor has been subjected to a speciation event, paralogs if it has been subjected to a duplication event and xenologs if they diverged via a HGT event.
Homologous sequences tend to have similar structure and function, and are often located in homologous genomic regions. These properties can be exploited in various biological applications, making deciphering the relation between genes essential for several biological analyses. For example, because homologous genes can be used as markers, they are essential in comparative genomics studies based on gene order, a field widely explored by renowned researchers in computational biology. In particular, Bernard Moret has led the development of highly efficient tools for comparing gene orders [5, 54, 55].
Methods for inferring gene relations are subdivided into tree-based and tree-free methods. Tree-free methods are mostly based on gene clustering according to sequence similarity, (cf., e.g., the COG database [87], OrthoMCL [50], InParanoid [10]). They are often unable to detect the full set of relations between members of a gene family and fail to differentiate orthologs from paralogs and xenologs. On the other hand, tree-based methods consist in reconstructing a phylogenetic tree for the gene family and then inferring the nature of internal nodes (duplication, speciation or HGT) from a reconciliation, i.e., an embedding of the gene tree into the species tree. Methods relying on reconciliation, the focus of this chapter, usually yield more accurate gene relations. However, they are very sensitive to the quality of the input trees, a single misplaced branch likely leading to a completely different evolutionary scenario.
Tree reconciliation can be performed through different biological models of evolution, the most common being the Duplication (D), Duplication-Loss (DL) or Duplication-Loss and Transfer (DTL) models. Incomplete lineage sorting (ILS), i.e., imperfect segregation of alleles has also been considered, mainly for reconciliation with a non-binary species tree. While most reconciliation methods are based on the parsimony principle of minimizing the number or the cost of induced operations, probabilistic models seeking for a reconciliation with maximum likelihood or maximum posterior probability have also been developed [2, 76, 84] (see [85] for a review). Although relying on more realistic models of gene family evolution through gains and losses, these methods are much slower than parsimony methods. This chapter is dedicated to parsimony methods for reconciliation.
As mentioned above, accurate inference of the true evolutionary history of a gene family through reconciliation strongly depends on the accuracy of the considered gene and species trees. This is the main reason for the continuing effort made to reduce errors in gene tree reconstruction. In particular, standard phylogenetic methods standing solely on sequence alignment (e.g., PhyML [33], RAxML [78], MrBayes [71], PhyloBayes [48]) are often error-prone as they are subject to, among other systematic errors [69], errors arising from the quality of the dataset (e.g., quality of gene annotations, gene family clustering, and alignment). In addition, gene sequences often do not contain enough differentiation (substitutions) to resolve all the branches of a phylogeny, or alternatively, too much such that the substitution history is saturated. The resulting low resolution of gene relations can usually be assessed with measures of statistical support (e.g., bootstrap and posterior probability) on tree branches.
To address the limitation of standard methods, other reconstruction methods, accounting for fitness with the species tree, have been developed. These methods, designated as integrative methods, report gene trees with better accuracy compared to sequence-only methods [14, 59, 84, 89]. Most of them rely on a two-steps approach: first compute a tree, or a set of trees, with the best fit to the sequences, and then “correct” the initial tree, or set of trees, according to the reconciliation cost. Four main strategies are considered for the second step: (1) Select neighboring gene trees of an initial tree by performing some branch swapping, typically Nearest Neighbor Interchange (NNI), Subtree Pruning and Regrafting (SPR) or Tree Bisection and Reconnection (TBR) (e.g., GeneTree [62], TreeFix [94], TreeFix-DTL [8], MowgliNNI [58], Notung [18]); (2) Contract branches of weak support and resolve the obtained polytomies (non-binary nodes) (e.g., NOTUNG [18], ProfileNJ [60]) ; Finally, select a set of trees or clades (leafsets) and construct (3) an amalgamated tree (e.g., ecceTERA [36], ALE [84] or (4) a supertree (e.g., MinSGT [41, 43]).
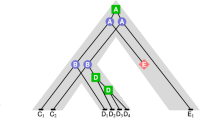
Different strategies for gene tree construction and correction. A single gene tree is constructed from the sequences of all the genes of the gene family: in (1), tree rearrangement methods around weakly supported branches are used to search an alternative tree minimizing with a better reconciliation cost; in (2), branches with weak support are rather contracted and the obtained non-binary nodes resolved according to the reconciliation cost with the species tree. (3) Amalgamation: a sample of gene trees is first reconstructed from a single gene family, then a single gene tree is reconstructed based on “trusted” clusters of the tree sample. (4) Supertree: The gene family is first subdivided into a set of, possibly overlapping, groups of genes (usually, groups of orthologs), a tree is reconstructed for each group and these trees are then combined into a single supertree displaying all of them
The first strategy, relying on tree rearrangement events (NNI, SPR, TBR) near poorly supported branches, consists of searching for alternative topologies of an initial gene tree with a better fit to the species tree. Methods based on this strategy explore the tree space often by using search heuristics such as branch-and-bound and hill-climbing. Some of them restrict the candidate alternative topologies to those that cannot be rejected by sequence data. Their main drawback stems from the performance of the criteria used to stop the tree space exploration, which in the worst case can result in exploring the complete exponential-size tree space.
In this chapter, while we focus on the second step of integrative methods, we only present the less straightforward methods based on strategies (2), (3) and (4). After introducing the preliminary notations in Sect. 5.2, the following sections are dedicated to the various formulations of the reconciliation problem depending on the considered trees and evolutionary model (with or without HGTs, considering or disregarding ILS). Section 5.3 is dedicated to the classical reconciliation between a binary gene tree and a binary species tree, Sect. 5.4 presents an extension to non-binary species trees, and Sect. 5.5 deals with the polytomy resolution problem, namely, the reconciliation of a non-binary gene tree with a binary species tree. This latter section is related to strategy (2) described above for integrative methods. We then move, in Sect. 5.6, to strategy (3) and (4), taking advantage of a set of gene trees rather than a single input gene tree, through amalgamation or supertree methods, as illustrated in Fig. 5.1. Section 5.7 then presents, for the DL model, a unifying view simultaneously considering polytomy resolution and supertree reconstruction in a single framework for gene tree correction. We end this chapter with a discussion in Sect. 5.8.
5.2 Trees
We denote, respectively, by V(T), E(T), and L(T) the set of nodes, edges and leaves of a tree T. Notice that \(L(T) \subset V(T)\). We say that T is a tree on L(T). Unless stated differently, all trees considered in this chapter are rooted, i.e., they admit a single node r(T) called the root of T.
Let x be a node of V(T); y is an ancestor of x if y is on the path from x to the root; y is a descendant (respectively, proper descendant) of x if y is on the path from x to a leaf of T including x (respectively, excluding x); x and y are incomparable if y is neither an ancestor nor a descendant of x. If (x, y) is an edge of T, then x is the parent p(y) of y and y is a child of x (\(y \in Ch(x)\)).
For a tree T, we denote by \(T_x\) the subtree of T rooted at \(x \in V(T)\). Two subtrees \(T_x\) and \(T_y\) of T are separated iff x and y are incomparable nodes of T. Given a subset L of leaves, we call the lowest common ancestor (LCA) of L in T and denote by \(lca_T(L)\) the common ancestor of L in T that is the farthest from the root. We also denote by \(T|_{L}\) the tree with leafset \(L \cap L(T)\) obtained from the subtree of T rooted at \(lca_T(L\cap L(T))\) by removing all leaves that are not in both L and L(T), and then all internal nodes with a single child.
A tree \(T'\) is said to be an extension of a tree T if it can be obtained by a sequence of graftings, where each grafting consists of subdividing an edge (x, y) of E(T) by creating a new node z between x and y, then adding a leaf l with parent z.
In this chapter, all considered trees have internal nodes with at least two descendants. An internal node x of T is binary if it has exactly two descendants. A binary tree is a tree with all internal nodes being binary nodes. A non-binary tree has at least one internal node which is a polytomy, i.e., a node with more than two descendants.
Definition 1
(binary refinement) A binary refinement \(B = B(T)\) of a tree T, is a binary tree such that \(V(T) \subseteq V(B)\) and such that for every \(x \in V(T)\), \(L(T_x) = L(B_x)\).
In other words, a binary tree B(T) is a binary refinement of T if whenever a node x is an ancestor of y in T, x is also an ancestor of y in B(G).
Gene and species trees: Two types of trees are considered: species trees and gene trees (see Fig. 5.2). A species tree S for a set \({\varSigma }= \{\sigma _1, \ldots , \sigma _t\}\) of species represents an ordered set of speciation events (the separation of one species into two different species) that have led to \({\varSigma }\).
Inside the species’ genomes, genes undergo speciation when the species to which they belong to speciate, but also duplication i.e., the creation of a new locus, loss of a locus, and Horizontal Gene Transfer (HGT) when a gene is transmitted from a source species to a different, coexisting target species.
A gene family \({\varGamma }\) is a set of genes sharing a common ancestor, and a gene tree G is a tree on a gene family \({\varGamma }\). We denote by s(x) the genome of \({\varSigma }\) to which x belongs.
When no distinction needs to be made between gene copies in the same genome, genes can just be identified by their corresponding genome, and thus a gene tree can be represented as a tree on \({\varSigma }\) with possibly repeated leaf-labels (see Fig. 5.3).
Top: A speciation (black circle), duplication (white rectangle), loss (dotted line) and HGT (white circle) events. For the speciation event, \(\sigma _l\) and \(\sigma _r\) refer to the two species descendent from the species \(\sigma \); for the HGT event, \(\sigma _1\) is the source and \(\sigma _2\) the target unrelated species. Bottom: (left) A gene tree G for the gene family \({\varGamma }= \{a_1, a_2, b_1, b_2, c_1\}\), where each lower case denotes a gene belonging to the corresponding genome in upper case; (middle) an evolutionary history of \({\varGamma }\) embedded in the species tree \(S = (A, (B,C))\); (right) the reconciliation R(G, S) corresponding to the given evolutionary history. Each internal node and grafted leaf x of R(G, S) is labeled with s(x). The edge \((B, a_2)\) is a HGT edge
5.3 Reconciliation of a Binary Gene Tree with a Binary Species Tree
The evolutionary history of a gene family is usually inferred from the embedding of its corresponding gene tree into the species tree, through a process called reconciliation explaining incongruities between gene and species trees by gene evolution events.
More precisely, a reconciliation R(G, S) of a gene tree G with a species tree S (if no ambiguity arises, we will just write R) is a node-labeled extension of the gene tree G reflecting a history of speciation and gene gain and loss in agreement with S (see Fig. 5.2). Each node x of V(R) (internal or leaf) is mapped to a node \(s(x) \in V(S)\). Some branches of R may also be labeled as transfer edges. A formal definition follows.
Definition 2
(Reconciled gene tree) Let G be a binary gene tree and S be a binary species tree. A reconciliation R(G, S) of G with S is an extension of G such that, for each internal node x of R(G, S) with two children \(x_l\) and \(x_r\), one of the following cases holds:
-
1.
\(s(x_l)\) and \(s(x_r)\) are the two children of s(x), in which case x is a speciation node;
-
2.
\(s(x_l) = s(x_r) = s(x)\) in which case x is a duplication node representing a duplication in s(x);
-
3.
one of \(s(x_l)\) and \(s(x_r)\) is equal to s(x) and the other is incomparable to s(x). Let y corresponds to the element of \(\{x_l,x_r\}\) such that s(y) is incomparable to s(x). Then x is a HGT node representing a HGT event with source genome s(x) and target genome s(y), and (x, y) is a HGT edge.
Each grafted leaf x corresponds to a loss in s(x).
Two genes are said orthologs if their LCA in R(G, S) is a speciation event, paralogs if it is a duplication event and xenologs if it is a HGT. For example in Fig. 5.2, \(b_2, c_1\) are orthologs, \(b_2, a_1\) are paralogs and \(a_2, b_1\) are xenologs.
Remark 1
A more flexible definition of xenologs, where two genes are said to be xenologs if the history since their LCA involves a HGT, is also considered in the literature [27]. With this definition, a pair of xenologous genes can diverge through speciation, duplication or transfer. For example with this definition, genes \(a_1, b_1\) in Fig. 5.2 are xenologs that diverged through a speciation. To avoid further ambiguity, a new classification of xenologs into subtypes, which takes into account the evolutionary events at the divergence of gene pairs and the relative timing of transfer and speciation events was also recently proposed [20]. In this chapter, we will consider the simplest event-based definition of xenologs through divergence via a transfer event, inducing a unique assignment type for each pair of genes into orthologs, paralogs or xenologs. Notice that with this definition, orthologs are not restricted anymore to genes from different species (see [20] for a discussion). For example, in Fig. 5.2, \(a_1\) and \(a_2\) are orthologs although they are found in the same present-day species A.
The standard parsimony criteria used to choose among the large set of possible reconciliations are the minimum number of duplications (D), duplications and losses (DL), or duplications, losses and HGTs (DTL) events induced by the reconciliation. The first two distances can be computed in linear time using the LCA mapping [30, 96, 99] (see Sect. 5.3.1 below). An algorithm enumerating all solutions for general costs with different event penalties was described in [22] for the DL model and extended to DTL in [15].
5.3.1 DL Reconciliation
The LCA-mapping between a gene tree G and a species tree S maps each node \(x \in V(G)\) toward a genome \(s(x) \in V(S)\), such as \(L(S_{s(x)})\) is the smallest set of genomes to which all genes in \(L(G_x)\) belong. Formally, \(s(x) = lca_S(\{s(y) : y \in L(G_x)\})\) in the species tree. Note that the LCA-mapping is unique for any given pair (G, S).
Given that mapping, each internal node x of G can be labeled as a duplication node if \(s(x_l)=s(x)\) and/or \(s(x_r)=s(x)\), otherwise it is a speciation node. The total number of losses correspond to the minimum number of grafting on G required to have a reconciliation R(G, S). The reconciliation induced by the LCA-mapping, called LCA-reconciliation is optimal for both D and DL distances. It is also the unique reconciliation minimizing the DL distance (see Fig. 5.3(1) for an example).
We highlight two types of duplication nodes inferred from LCA mapping. Consider each gene of G as simply identified by the genome it belongs to. Let x be a duplication node of G with children \(x_l\) and \(x_r\). It is a Nonapparent duplication (NAD) iff \(L(G_{x_l})\cap L(G_{x_r}) = \emptyset \). In other words, the reason for x being a duplication node is not the presence of paralogs in the same genome, but rather an inconsistency with the species tree. A duplication which is not a NAD is an Apparent Duplication (AD) node, i.e., a node with the left and right subtrees sharing a common leaf-label.
For example in Fig. 5.3(1), the lower duplication node of G is a NAD, while the upper duplication node is an apparent duplication, as its left and right subtrees each contains a leaf labeled b.
While apparent duplications are supported by the presence of paralogs, in the same genome, that are necessarily the result of duplication, NAD nodes have been flagged as potential errors in many studies, and in particular in the Ensembl Compara gene tree database [28]. The distinction between these two types of duplication nodes is required for certain formulations of the gene tree correction problem [40], or for considering an optimal history accounting for ILS, as we will see later.
Three different reconciliations for the species tree S and the gene tree G, for the gene family \({\varGamma }=\{a,b,b,c,d\}\), where each lower case denotes a gene belonging to the corresponding genome in upper case; (1) An evolutionary scenario optimal for the D and DL distances (two duplications and five losses); G is labeled according to the LCA-mapping; (2) A DTL-scenario with two HGTs and two losses. This scenario is cyclic, and is therefore infeasible; (3) An alternative and acyclic DTL-scenario with two HGTs and one loss; this scenario is also biologically unfeasible as it is not date-respecting, according to the considered speciation times
5.3.2 DTL Reconciliation
In contrast with the DL reconciliation framework, the optimal DTL reconciliation is not unique, and cannot be computed by means of the LCA-mapping. With HGTs, a gene evolution is not restricted anymore within the parental edges of its genome in the species tree. As such, to the standard vertical transmission of genes from one ancestor genome to its descendants, there is an additional need to consider transmission between incomparable nodes of the species tree. Such transmissions are represented in the reconciliation by a transfer edge (x, y) corresponding to a gene transfer from a source genome s(x) to a target genome s(y). For a HGT to be biologically feasible, both genomes are required to be contemporary at the time of the transfer event. Therefore, a “consistent” HGT scenario should allow a total temporal ordering of the internal nodes of the species tree S. As demonstrated by Tofigh et al. [91], this requires the DTL-reconciliation to be acyclic, as defined below.
Definition 3
A reconciliation R(G, S) is acyclic if and only if there is a total order < on V(S) such that:
-
(1)
if \((s,s') \in E(S)\) then \(s<s'\) and
-
(2)
if (x, y) and \((x',y')\) are transfer edges in G such that \(y'\) is a descendant of y in R(G, S), then \(p(s(x)) < s(y')\).
For example, scenario 2 in Fig. 5.3(2) is a cyclic DTL-scenario, as the ordering defined by the above definition would lead, for the two transfer edges of G, to \(\alpha < \alpha \). On the other hand, scenario 3 (Fig. 5.3(3)) is acyclic.
The problem of finding a most parsimonious acyclic (i.e., time-consistent) DTL-scenario is NP-hard [23, 24, 34, 61]. However, the problem becomes polynomial if the acyclicity requirement is dropped [6, 91]. In that case, the main idea for computing an optimal DTL-reconciliation is to consider all possible mappings of G nodes to S nodes, using a dynamic programming approach.
More precisely, let c(x, s) be the minimum cost of a reconciliation of \(G_x\) with S such that x is mapped to \(s \in V(S)\). The gene tree G is processed in post-order traversal, with the base case corresponding to leaves \(x \in L(G)\), treated as follows:
As for an internal node x with children y and z, we have to consider the three possibilities of x being labeled as a speciation, duplication or HGT node, with \(c_s(x,s)\), \(c_d(x,s)\), and \(c_t(x,s)\) representing these three mutually exclusive cases. Then, \(c(x,s) = \min \{c_s(x,s),c_d(x,s),c_t(x,s)\}\). Finally, the minimum cost of a reconciliation of G with S is \(\min _{s \in V(S)} c(r(G),s)\).
For simplicity, we report below the recurrences when considering the cost of reconciliation as being the number of duplications and HGT [91].
A straightforward implementation of these recurrences lead to an algorithm in \(O(mn^2)\) time, where \(m = |V(G)|\) and \(n = |V(S)|\). This time complexity has been further improved to O(mn) [90].
Notice that losses may be essential for distinguishing between duplications and HGT events. The above recurrences have to be adapted to handle losses. David and Alm [21] have described an algorithm for the DTL distance running in \(O(mn^2)\), while Bansal et al. [6] described RANGER-DTL, an algorithm running in O(mn).
When divergence time information, or a temporal ordering of internal nodes, is available for S, then the DTL-scenario must respect this ordering (i.e., HGT events are constrained to occur only between coexisting species). A DTL-scenario respecting a dated tree is called a date-respecting DTL-scenario. Bansal et al. [6] show how the definition of a reconciliation and the above recurrences can be adapted to solve this problem. They give an algorithm with \(O(mn \log n)\) time complexity.
For example, scenario 3 of Fig. 5.3 is not date-respecting. Notice that a date-respecting DTL-scenario is not necessarily time-consistent. In fact, scenarios may be locally consistent (i.e., HGT events occurring between coexisting species), but globally inconsistent. Global consistency may be obtained by subdividing the species tree S into slices and exploring slices one after the other. This strategy has been first used in [51], leading to an \(O(nm^4)\) algorithm. Later, Doyon et al. [24] have improved the computation of a most parsimonious time-consistent DTL-reconciliation with a dated species tree to \(O(mn^2)\).
5.3.3 Binary Gene Tree Reconciliation in Presence of ILS
When a population of individuals undergoes a series of speciations in a short period of time, different alleles for the same gene locus may remain present in a given lineage, and then eventually fixed differently in descendant lineages [52]. This phenomenon, known as deep coalescence or Incomplete Lineage Sorting (ILS) may also explain discrepancies between a gene tree and a species tree. For example in Fig. 5.4, the subtree \(((a,b_1),c_1)\) of G, which is incongruent with the species tree (A, (B, C)), may be explained from the history depicted in the left backbone of (i), which involves no duplication, but simply the fact that the allele inherited in C is different from the one inherited in A and B.
Gene family evolution and incomplete lineage sorting. (i) Evolution of a gene family inside a species tree \(S = (A,(B,C))\), in the context of a population. Each species tree backbone contains the evolution of a single locus and each row represents a generation of individuals in a population. The lines inside the tree backbones represent the evolution of the gene family leading to the tree G in (ii). In this example, the evolution of two loci (black and green) are depicted. Two alleles of the black locus are present at the time of speciation 1. The first allele is fixed in A and B, whereas the second is only fixed in C. The green locus was created after an ancestral duplication occurring just before speciation 1, and was lost in genome A; (ii) The resulting gene tree G for the gene family \({\varGamma }= \{a, b_1, b_2, c_1, c_2\}\) is the represented reconciled tree R(G, S), ignoring losses (dotted lines) and internal node labeling. Duplication nodes, inferred from the LCA-mapping, are not coherent with the true evolutionary history of the gene family. (iii) A different representation of R(G, S) reflecting the number \(n_s\) of gene copies in each genome s. For example, for the branch (0, 1), we have \(n_0 = 1\) and \(n_1 = 3\). (iv) A different scenario able to explain incongruities between the gene and species tree through duplication, loss and deep coalescence. This more parsimonious history involves one duplication, a loss, and a deep coalescence event. It relies on the labeled coalescent tree model which simultaneously describes the species, locus and gene trees, as well as the reconciliations between them. (v) The locus tree (LT) induced by the scenario shown in (iv). (vi) Enumeration of the possible locus maps for each branch of the species tree. Each locus is shown with a different color and new locus are created by duplications. Only some locus maps for branches (0, 1) and (1, A) are shown. The mapping is based on the total number of gene lineages at the start and end of each edge of the species tree, which can be determined with LCA-mapping
In the absence of paralogous genes in the same genome, inconsistencies between a gene tree and a species tree can always be explained through ILS. Wu and Zhang [93] have shown that a unique reconciliation with minimum deep coalescence cost can be obtained in that case, using LCA-mapping. It is, however, necessary to take duplication events into account as ILS cannot explain the presence of additional loci. For example, in Fig. 5.4, while the NAD (nonapparent duplication) in G can be adequately explained through ILS, the apparent duplication node above it necessarily involves the creation of a second locus. As seen in Fig. 5.4(iii), (iv), ILS-aware reconciliation methods may produce evolutionary histories with fewer losses, highlighting the need of models jointly considering duplication, loss, HGT and ILS events. In a recent paper, Bork et al. [13] have shown that the duplication-loss-ILS reconciliation problem is NP-hard, even when only duplications are to be minimized.
Very few papers have attempted to jointly model ILS and other macro-evolutionary events during gene and species tree reconciliation. In two papers by Durand’s group [81, 92], the problem is reformulated as a reconciliation between a binary gene tree and a non-binary species tree minimizing the DL/DTL cost. Their algorithm first requires contraction of short branches of the species tree into polytomies and ILS are only allowed at those unresolved nodes and remain unpenalized. Section 5.4 is dedicated to this algorithm.
On the other hand, Kellis et al. [67, 95] have considered a coalescent model for reconciling a binary gene tree with a binary species tree, accounting for duplications, losses and deep coalescence. They first devised a probabilistic algorithm, called DLCoal [67]. Although efficient, this algorithm is highly parameterized, making it impracticable. Subsequently, they proposed a parsimony-based algorithm, called DLCpar [95], introducing the concept of a label coalescent tree (LCT) (see Fig. 5.4(iv)), which simultaneously describes the reconciliation between a gene tree, a locus tree, and a species tree. This latter algorithm proceeds in the following steps:
-
1.
Use the LCA-mapping between G and S to determine all implied speciation nodes and count, for each branch (x, y) of the species tree, the numbers \(n_x\) and \(n_y\) of gene copies at x and y.
-
2.
For each branch (x, y), in a pre-order traversal of S, enumerate all possible scenarios of DL and ILS events leading from \(n_x\) to \(n_y\) gene copies (see Fig. 5.4(vi)). This yields the set of possible species-specific locus maps that associates each node of the gene tree to the locus in which it evolves. The event cost for each branch of S can be computed by counting the number of additional loci and lost loci, respectively corresponding to duplications and losses, as well as the number of extra lineages caused by deep coalescence (see Fig. 5.4(vi)). In practice, some histories are not considered since they are never most parsimonious.
-
3.
Perform a post-order traversal of S, and for each branch (x, y) and each assignment \((n_x, n_y)\), use dynamic programming to determine the minimum cost on the subtree of S rooted as x, computed as the cost of the branch (x, y) plus the minimum cost of the left and right subtrees rooted at y, where y is assigned \(n_y\) loci. The minimum among all possible choices is selected as the most parsimonious reconciliation. Optimal loci at the start and end of each branch can then be assigned with a traceback starting from the root of the species tree.
Although not explicitly given in the paper, the complexity of the algorithm strongly depends on the size of the locus maps set and on the choices considered for each branch of the species tree. This part is not detailed in the paper. In particular, the method is supposed to search over the entire space of reconciliations, but it is not clear whether it leads to a heuristic or to an exact algorithm.
In a follow-up paper, Rogers et al. [70] further attempt to extend the LCT model in order to address one of its shortcomings, namely the assumption of a single haploid sample for each species. More recently, Chan et al. [16] have proposed the first FPT (fixed-parameter-tractable) algorithm that computes the most parsimonious time-consistent reconciliation fully accounting for ILS, duplications, HGTs and losses (IDTL). This algorithm is an extension of the DTL-reconciliation described in [24] with modifications to allow ILS, and has a total complexity of \(O(|V_G|(|V_S|^2 + |V_S|n_k{2^{k_S}})2^k_s)\), where k is the number of branches in the largest ILS subtree (i.e., subtrees of the species tree where ILS occur) and \(n_k\) the number of ILS subtrees.
5.4 Reconciliation with a Non-binary Species Tree
The LCA-mapping can naturally be generalized to a non-binary species tree. However, the LCA-reconciliation used for binary gene and species trees will not produce correct gene evolution history when applied to non-binary species trees. In fact, a node of G and its child mapping to the same non-binary node of the species tree does not necessarily indicate a duplication. In [97], Zheng et al. proved that the general reconciliation problem of a gene tree G with a non-binary species tree S via binary refinement is NP-hard, even when only duplications are considered. In the same paper, they proposed a heuristic for the problem also allowing for polytomies in the gene tree.
We can distinguish two reasons for the presence of non-binary nodes in a species tree. They can either represent “true” evolutionary events, i.e., adaptive radiations leading to the emergence of a set of species from a single ancestral one, or can be caused by a lack of resolution in the species tree, due to methodological reasons. Such non-binary nodes are called hard in the former case and soft in the latter case. A soft polytomy may be due to short time since speciation, leading to genetic drift.
In either case, non-binary nodes of a species tree often correspond to populations with substantial genetic diversity, and coexisting multiple alleles. It is expected that some gene families might exhibit imperfect segregation of all their alleles (in other words ILS) at these nodes. Therefore, a subtree of the gene tree whose root maps to a polytomy in the species tree may be differently explained by speciation, duplication or ILS, depending on the considered resolution of that polytomy.
Vernot et al. [92] have considered the problem of finding a most parsimonious DL scenario explaining the differences between a binary gene tree G and a non-binary species tree S, assuming that disagreements between the two trees can stem from either duplication or ILS. Their algorithm only considers the possibility of ILS at non-binary nodes of S. The main idea of their algorithm is to identify required duplications, i.e., those disagreements with the species tree that can only be explained by a duplication. Clearly, these nodes are those in G that would be labeled as duplication in all resolutions of S. However, as shown in [92], there is no need to try all the resolutions of S.
The procedure described in [92] consists of a post-traversal of G during which each node x of \(V(G)\setminus \{r(G)\}\) is labeled by the set N(x), which is the subset of \(\{h: h \in Ch(s(p(x)))\}\) such that each element \(h \in N(x)\) has at least one descendant in \(\{s(l): l\in L(G_x)\}\). This set represents the minimum set of nodes in V(S) that would be traversed from s(x) to the mapping of x’s children, regardless of the resolution of S. Consequently, a node x with children \(x_l\) and \(x_r\) is a required duplication if and only if \(N(x_r) \cap N(x_l) \ne \emptyset \) (see Fig. 5.5 for an example).
A species tree S for the genome set \({\varSigma }= \{A,B,C,D\}\); A gene tree G for the gene family \({\varGamma }=\{a,a,b,c,d\}\), where each small letter designs a gene belonging to the corresponding genome in upper case. The tree G is labeled according to LCA-mapping suggesting three duplication nodes (rectangles). However, according to the N(x) labeling in brackets, only two duplications are required, while the third (striped rectangle/circle) can be explained through ILS instead (see history in the right side), leading to a most parsimonious DL scenario involving two duplications and four losses
The set labeling a node of G is of size \(O(k_S)\) where \(k_S\) is the maximum outdegree in S. Based on this fact, Vernot et al. [92] have described an algorithm for the D distance running in \(O(|V(G)|(k_S + h_S))\) time, where \(h_S\) is the height of S (i.e., maximum number of nodes from the root to any leaf of S). However, inferring the induced minimum number of losses is not as straightforward as for binary species trees. In fact, for a loss associated to a polytomy, it is not generally possible to determine the exact lineage in the gene tree in which the loss has occurred, and several edges of G have to be tested. An exponential algorithm running in \(O(|V(G)|k_S2^{2k_S})\) was described.
In [81], Stolzer et al. further extended the framework to HGT events and developed an algorithm running in \(O(|V(G)|(h_S + k_S)(V|S| + n_k2^{k_S} )^2)\). Although their algorithm does not guarantee a time-consistent reconciliation, temporal feasibility of each scenario is evaluated a posteriori. Both DL and DTL algorithms are implemented in NOTUNG.
5.5 Reconciliation of a Non-binary Gene Tree with a Binary Species Tree
We will detail the most efficient algorithms for DL reconciliation, and end up with a brief discussion on extensions to DTL reconciliation of a non-binary gene tree G with a binary species tree S. This problem is motivated by the gene tree correction problem, where a non-binary gene tree can be obtained from an initial tree by contracting weakly supported branches. In other words, the polytomies of G are considered soft, i.e., reflect non-resolved parts of the tree. The goal is then to find an appropriate refinement (as defined in Sect. 5.2) of this non-binary gene tree.
Definition 4
(Resolution) A resolution of G with respect to S is a reconciliation R(B, S) between a binary refinement B of G and S. The set of all possible resolutions of a gene tree G is denoted \({\mathscr {R}}(G)\).
The optimization problem follows.
Minimum Resolution Problem:
Input: A binary species tree S and a non-binary gene tree G.
Output: A Minimum Resolution of G with respect to S (or simply Minimum Resolution of G), e.g., a resolution of G of minimum reconciliation cost with respect to S.
As first noticed by Chang and Eulenstein [17], each polytomy of G can be considered independently and a minimum resolution of G can be obtained by a depth-first procedure that iteratively solves each polytomy \(G_x\) for each internal node x of G.
An \(O\big (|V(S)||V(G)|^3\big )\) algorithm for the resolution of a non-binary gene tree minimizing duplications and losses was first considered in NOTUNG [25]. The same year, Chang and Eulenstein [17] also described an algorithm with a better complexity, running in \(O\big (|V(S)||V(G)|^2\big )\). In 2012 [45], we developed the first linear-time algorithm for resolving a polytomy (a single unresolved node), leading to an overall quadratic-time algorithm for a whole tree. An algorithmic result extending linearity to a whole gene tree was later obtained by Zheng and Zhang [98]. The key idea is to resolve each polytomy with a species tree restricted to the smallest necessary set of genomes. Their algorithm does not allow, however, to output all solutions and is restricted to unit cost for duplications and losses. Based on the same optimization idea, we developed PolytomySolver [42] which is a generalization of the dynamic programming algorithm given in [45], allowing for both event-specific and species-specific costs. The time complexity of PolytomySolver is linear for the unit cost and quadratic for the general cost, which outperforms the best-known solutions so far by a linear factor.
In the rest of this section, we describe the dynamic programming technique in PolytomySolver for the resolution of a single polytomy under the DL distance with unitary event costs. More details, complexity improvement, extension to other costs and to a full non-binary gene tree, can be found in [42].
5.5.1 PolytomySolver
In the following, to prevent penalizing losses in genomes with no descendant genes in G, the species tree is restricted to \(S|_{\{s(x)\; :\; x\in L(G) \}}\) and we will simply continue to refer to it as S.
(Figure from [42]; use permitted under the Creative Commons Attribution License CC-BY 3.0) A polytomy G and a species tree S. Squares on trees illustrate duplications, whereas speciation are denoted by a black circle. To the right of table M, the forests corresponding to an (a, 1) and (a, 3)-resolution are given, where the gray circled a illustrates a loss. We illustrate the (d, 1)-resolution, rooted at a speciation node, corresponding to \(C_{d,1} =3\) (obtained from the vertical arrows in table M), and an optimal (d, 1)-resolution, obtained from a (d, 2)-resolution (horizontal arrow in M). The optimal cost for the resolution of G (\(M_{e,2} =2\)) is highlighted in blue
PolytomySolver proceeds with a recursion made on the subtrees of S. Define the multiplicity m(s) of \(s \in V(S)\) in G as the number of times it appears in G, i.e., \(m(s) = |\{x \in L(G) : s(x) = s\}|\). An (s, k)-resolution of G is a forest of k reconciled gene trees \({\mathscr {T}}= \{T_1, \ldots , T_k\}\) s.t. \(\forall \; 1 \le i \le k\), \(s(r(T_i)) = s\), and each leaf x of G with s(x) being a descendant of s is present as a leaf of some tree of \({\mathscr {T}}\) (see Fig. 5.6 for an example). Leaves of trees in \({\mathscr {T}}\) that do not appear in L(G) represent losses. We denote by \(c({\mathscr {T}})\) the reconciliation cost of the forest \({\mathscr {T}}\). This cost is the sum of the reconciliation costs of all \(T_i \in {\mathscr {T}}\).
The cost of a minimum resolution of G can be computed using a dynamic programming algorithm that fills a table M. Each cell \( M_{s, k}\) of M corresponds to the minimum cost of an (s, k)-resolution for a given node s of S and a given integer \(k \ge 1\) (\(M_{s, k} = +\infty \) for \(k < 1\)). The final cost of a minimum resolution of G is given by \(M_{r(S),1}\). The table M can be computed, line by line, in a bottom-up traversal of S. Although k is unlimited (number of gene losses is unlimited), we have shown in [42] that there is no need to consider values larger than \(|V(G)| - 1\).
Lemma 1 gives the base case to compute \(M_{s,k}\) when \(s \in L(S)\). It follows from the fact that, if k is larger than the number of available leaves, then additional leaves corresponding to gene losses are required; otherwise, leaves have to be joined under duplication nodes. An illustration of this lemma is shown in Fig. 5.6 where it is used to compute the first three lines of M.
Lemma 1
(Base case) For a leaf node s of S, if \(k > m(s)\) then \(M_{s,k} = k - m(s)\); otherwise \(M_{s,k} = m(s) - k\).
For an internal node s of S, speciation events also need to be considered. We require an intermediate cost table C where each entry \(C_{s, k}\) represents the minimum cost of an (s, k)-resolution in which every tree is rooted at a speciation node with two children or is a leaf of G already mapped to s. For \(k > m(s)\), an (s, k)-resolution of cost \(C_{s, k}\) can only be obtained from an \((s_l, k - m(s))\)-resolution and an \((s_r, k - m(s))\)-resolution by first generating \(k - m(s)\) speciation nodes, mapped to s, each joining a pair \((s_l, s_r)\), then adding the m(s) trees already available (see for example the (d, 1)-resolution corresponding to \(C_{d,1}\) in Fig. 5.6; in this case \(m(d)=0\)). Thus, we define:
As nodes mapped to s are not necessarily speciation nodes but can also correspond to duplications, it is readily seen that \(M_{s, k} \le C_{s, k}\). A recurrence for computing \(M_{s,k}\) follows.
Lemma 2
For an internal node s of S, \(M_{s,k} = \min (M_{s,k - 1} + 1, M_{s, k + 1} + 1, C_{s, k})\).
In Lemma 2, the first term of \(M_{s,k}\) corresponds to a loss, while the second corresponds to a duplication at s.
Since \(M_{s, k}\) depends on \(M_{s, k + 1}\) and vice-versa, the recurrence cannot be used to compute C and M. This dependency can, however, be avoided due to a strong property on lines of M. In [45] we have shown that each line \(M_s\) is characterized by two values \(k_1\) and \(k_2\) such that, for any \(k_1 \le k \le k_2\), all \(M_{s, k}\) have a single minimum value \(\gamma \), for any \(k \le k_1\), \(M_{s,k-1} = M_{s,k}+1\), and for any \(k \ge k_2\), \(M_{s,k+1} = M_{s, k}+1\). In other words, \(M_s\) can be treated as a convex function fully determined by \(k_1, k_2\) and its minimum value \(\gamma \). We say \(M_s\) has a minimum plateau between \(k_1\) and \(k_2\). For example, line \(M_d\) in Fig. 5.6 is fully determined by \(k_1 = 2\) and \(k_2 = 3\) and its minimum value \(\gamma _d = 1\).
Theorem 1
(Recurrence 1) Let \(k_1\) and \(k_2\) be the smallest and largest values, respectively, such that \(C_{s, k_1} = C_{s, k_2} = \min _{k} C_{s, k}\). Then,
Theorem 1 shows how a row \(M_s\) for an internal node s of S can be computed: for each k, compute \(C_{s, k}\) using recurrence Theorem 1 and keep the two columns \(k_1\) and \(k_2\) setting the bounds of the convex function’s plateau. The \(M_{s,k}\) values at the left and right of the minimum plateau can then be easily computed from the value of the minimum plateau. These recurrences, with the base case for S leaves given in Lemma 1, describe how the dynamic programming algorithm of PolytomySolver works.
Algorithm 1 describes the computation of table M. We refer the reader to [45] for the reconstruction of a solution from M, which is accomplished using a standard backtracking procedure. Moreover, we show in [42] that \(k_1\) and \(k_2\) for each M(s) can be computed in constant time from \(M_{s_l}\) and \(M_{s_r}\) vectors. This implies a linear-time algorithm for the computation of \(M_{root(S),k}\).

Unrooted trees: If the gene tree is unrooted, an exhaustive testing of all roots can be done with PolytomySolver, ProfileNJ [60] and NOTUNG [18]. A series of papers by Gorecki et al. also consider the properties of the plateau to avoid exploring all branches [31, 32] of unrooted gene trees.
5.5.2 Extensions to DTL Reconciliation
The dated and undated formulations of the DTL reconciliation have been shown to be NP-hard for non-binary gene trees [38]. Kordi and Bansal [39] have also shown that the problem is Fixed-Parameter-Tractable (FPT) in the maximum degree k of the gene tree, and explored a \(O\big (2^kk^k(|V(S)|+|V(G)|)^{o(1)}\big )\) algorithm testing all possible resolutions of the gene tree. A similar algorithm, implemented in NOTUNG [47], also tries all possible resolutions of each polytomy before computing the DTL distance for each resolution. Heuristics for the problem, including exploration of the tree space surrounding an initial resolution were also implemented in NOTUNG. One such possibility consists of selecting a best tree for the DL reconciliation, and then exploring alternative topologies at a given maximum NNI distance from the initial topology. Finally, Jacox et al. [37] have also proposed an algorithm improving the time complexity to \(O\big ((3^k-2^{k+1})(V(|S|)+V(|G|))^{o(1)}\big )\) by using amalgamation principles (see Sect. 5.6). Although this algorithm improves the running time by an exponential factor, it runs in \(O(2^k)\) space compared to the algorithm described in [39] requiring polynomial space complexity.
5.6 Inferring a Gene Tree from a Set of Trees
We now move to a slightly different gene tree correction strategy, which consists of taking advantage of a set of gene trees rather than a single input gene tree.
5.6.1 Amalgamation: Gene Tree Inference from a Set of Clades
As sequence information may contain limited signal, phylogenetic reconstruction often involves choosing among a set of equally likely trees. This idea has inspired the amalgamation procedure for reconstructing a tree from the clades, i.e., subtree leafsets, of a set of gene trees. This principle was first introduced by David and Alm [21] and a heuristic for correcting an initial gene tree based on this idea has been described. The amalgamation principle was extended by Szöllősi et al. [84] in a probabilistic method called ALE (for Amalgamated Likelihood Estimation) considering conditional clade probabilities (introduced in [35]) and a joint sequence-reconciliation likelihood score.
An alternative deterministic algorithm, called TERA (for Tree Estimation using Reconciliation) has been developed by Scornavacca et al. [74]. This algorithm “amalgamates” the most parsimonious DTL reconciled gene tree from an initial set of gene trees and achieves similar accuracy than ALE, while being much faster.
We start with some definitions, before presenting the outline of TERA.
Definition 5
Given a tree T and a node x of T, we call \(L(T_x)\) the clade of T at x and denote by \({\mathscr {C}}(T)\) the set of all clades of T. If x is an internal node with children \(x_l\) and \(x_r\), a tripartition at x is defined as \(\pi _x = (\pi _x[1],\pi _x[2],\pi _x[3])\) with \(\pi _x[1] = L(T_x)\), \(\pi _x[2] = L(T_{x_l})\) and \(\pi _x[3] = L(T_{x_r})\). Given a set \(\mathcal{G}\) of k gene trees on the same gene family \({\varGamma }\), we denote by \({\mathscr {C}}(\mathcal{G})\) the set of all the clades of \(\mathcal{G}\), and by \(\Pi (\mathcal{G})\) the union of all tripartitions of \(\mathcal{G}\). For a given clade \(c\in {\mathscr {C}}(\mathcal{G})\), \(\Pi (c)\) corresponds to the set of tripartitions \(\pi \) of \(\Pi (\mathcal{G})\) such that \(\pi [1]=c\).
Definition 6
(Amalgamation) An amalgamation of \(\mathcal{G}\) is any gene tree G on \({\varGamma }\) such that \({\mathscr {C}}(G) \subset {\mathscr {C}}(\mathcal{G})\).
Most Parsimonious Amalgamation problem
Input: A set \(\mathcal{G}\) of gene trees on the gene family \({\varGamma }\), and \({\mathscr {C}}(\mathcal{G})\) the set of all the clades of \(\mathcal{G}\).
Output: An amalgamation of \(\mathcal{G}\) minimizing the reconciliation cost with respect to S.
The TERA algorithm solves the amalgamation problem by computing the optimal reconciliation of each clade (i.e., polytomy with clade as leafset) with each node of S. For that purpose, the algorithm performs a joint traversal of the species tree S and the clades of \({\mathscr {C}}(\mathcal{G})\). In an initial step, it computes the reconciliation of each clade \(c \in {\mathscr {C}}(\mathcal{G})\) with the leaves of S. Then S is traversed bottom-up, and for each node \(s \in V(S)\), the reconciliation cost of each tripartition of c with s is computed. For each pair (c, s), the algorithm computes the cost of reconciling the clade c with s by testing all possible tripartitions \(\pi \) in \(\Pi (c)\). As each non-trivial tripartition \(\pi \) can be seen as an internal node of an amalgamated tree with children \(\pi [2]\) and \(\pi [3]\), the cost of reconciling a tripartition \(\pi \) with s can be computed, using the recurrences of the DTL-reconciliation algorithm [24] (see Sect. 5.3), from the cost of reconciling \(\pi [2]\) and \(\pi [3]\) respectively with nodes of \(V(S_s)\). The output of TERA is the most parsimonious reconciliation at one of the root clades.
The TERA algorithm is part of a unifying software called ecceTERA [36] accounting for a variety of evolutionary events including duplications, losses, transfers, transfer-loss and transfers from/to an unsampled species (not represented by the set of genes). The software also handles fully or partially dated, as well as undated, species trees.
5.6.2 Supertree: Inferring a Tree from a Set of Subtrees
Homology-based search tools are usually used to seek all homologs of a given gene in a set of genomes. The resulting gene family may be very large, involving distant gene sequences that may be hard to align, leading to weakly supported trees. Alternatively, gene copies may be grouped into smaller sets of orthologs and inparalogs, using clustering algorithms such as OrthoMCL [50], InParanoid [10], Proteinortho [49] or many others.Footnote 1 Trees obtained for such partial gene families should then be combined into a single one using a supertree method.
Supertree methods have been mainly designed to reconstruct a species tree from gene trees obtained for various gene families (see for example [7, 11, 57, 64, 65, 72, 80, 83]). However, they can have applications for gene tree reconstruction as well. In this case, a gene tree is constructed from a set of subtrees for partial, possibly overlapping, subsets of the gene family. Ideally, the obtained tree should display each of the input trees, which is only possible if the partial trees are consistent, i.e., exhibit the same topology for each triplet of genomes (assuming genes are simply represented by the genome they belong to).
The simplest formulation of the supertree problem is therefore to state whether an input set of trees is consistent, and if so, find a compatible tree, called a supertree, displaying them all. This problem is NP-complete for unrooted trees [73, 79], but solvable in polynomial time for rooted trees [1, 19, 56, 75]. The BUILD algorithm [1] can be used to test, in polynomial time, whether a collection of rooted trees is consistent, and if so, construct a compatible, not necessarily fully resolved, supertree. This algorithm has been generalized to output all supertrees [19, 56, 75], which may be exponential in the number of genes.
Supertree methods can also be used to correct gene trees, by removing weakly supported upper branches and then constructing a supertree from the set of terminal subtrees. In contrast with the polytomy resolution approach, neither the input subtrees, nor the gene clusters of those subtrees are necessarily preserved. In other words, the exhibited monophyly of input gene clusters can be challenged. This is particularly relevant because it has been shown that genes under negative selection, while exhibiting the true topology, might be wrongly grouped into monophyletic groups (see for example [53, 77, 82, 88]). Using a supertree method might, therefore, be beneficial, as it preserves the topology of subtrees, while allowing to group genes from different subtrees.
In [41, 43], we introduced the MinSGT problem defined as follows.
Minimum SuperGeneTree (MinSGT) Problem:
Input: A species set \({\varSigma }\) and a species tree S for \({\varSigma }\); a gene family \({\varGamma }\) of size n, a set \({\varGamma }_{i, 1 \le i \le k}\) of potentially intersecting subsets of \({\varGamma }\) such that \(\bigcup _{i=1}^{k}{{\varGamma }_i} = {\varGamma }\), and a consistent set \({\mathscr {G}}= \{G_1, G_2,\ldots , G_k \}\) of gene trees such that, for each \(1 \le i \le k\), \(G_i\) is a tree for \({\varGamma }_i\).
Output: Among all trees G for \({\varGamma }\) and compatible with \({\mathscr {G}}\), one of minimum reconciliation cost.
Under the D distance, we have shown that this problem is NP-hard to approximate within a \(n^{1 - \varepsilon }\) factor, for any \(0< \varepsilon < 1\), even for instances in which there is only one gene per species in the input trees, and even if each gene appears in at most one input tree. Although it has not been proven yet, MinSGT is conjectured NP-hard for the DL reconciliation cost, as accounting for losses in addition to duplications is unlikely to make the problem simpler.
We developed a dynamic programming algorithm for MinSGT with the DL reconciliation cost, which has a time complexity exponential in the number of input trees. The algorithm constructs the supertree G from the root to the leaves. At each step, i.e., for each internal node x being constructed in G, all possible bipartitions \((B_l(x),B_r(x))\) that could be induced by x are tried, and the iteration continues on each of \(B_l(x)\) and \(B_r(x)\). The goal is to find the bipartition of \({\varGamma }\), that leads to the minimum DL reconciliation cost at the root. At each step, corresponding to a node x, the reconciliation cost is computed from a local reconciliation cost at x, and from the best reconciliation cost of the two clusters of the considered bipartition. Because of the constraint of being compatible with the input gene trees only a subset of the bipartition set need to be tested at each step.
Property 1
Let \({\mathscr {G}}= \{G_1,\ldots ,G_k\}\) be a set of gene trees. The root of a supertree G compatible with \({\mathscr {G}}\) subdivides \(\bigcup _{i=1}^{k}{L(G_i)}\) into a compatible bipartition \((B_l,B_r)\), i.e., a bipartition such that, for each i s.t. \(1 \le i \le k\), either: (1) \(L(G_i) \subseteq B_l\); or (2) \(L(G_i) \subseteq B_r\); or (3) \(L(G_{i_l}) \subseteq B_l\) and \(L(G_{i_r}) \subseteq B_r\); or (4) \(L(G_{i_l}) \subseteq B_r\) and \(L(G_{i_r}) \subseteq B_l\).
An illustration of the seven valid bipartitions for two trees \(G_1\) and \(G_2\). Each bipartition is obtained by “sending” \(L_1 \in \{L(G_1), L(G_{1, l}), L(G_{1, r}), \emptyset \}\) in the left part, and the complement \(L(G_1) \setminus L_1\) in the right part. The same process is then applied to \(G_2\). The set \(\mathscr {B}(G_1, G_2)\) consists of the set of all possible combinations of choices, after eliminating symmetric cases and partitions with an empty side
Let \(\mathscr {B}(G_1, \ldots , G_k)\) be the set of all possible combinations of choices resulting from Property 1 (see Fig. 5.7 for an example). Notice that not all such combinations are valid bipartitions. For instance in Fig. 5.7, the first bipartition (top-left) cannot be valid if \(G_1\) and \(G_2\) share a leaf with the same label, as a gene cannot be sent both left and right. These cases, however, can be detected easily by verifying the leafset of \(B_l\) and \(B_r\).
Denote by \(MinSGT(G_1,\ldots ,G_k)\) the minimum DL reconciliation cost of a supertree compatible with \({\mathscr {G}}= \{G_1,\ldots ,G_k\}\). The main recurrence formula of the dynamic programming algorithm is stated as follows.
Theorem 2
Let \({\mathscr {G}}= \{G_1,\ldots ,G_k\}\) be a set of gene trees.
-
1.
\(MinSGT(G_1,\ldots ,G_k) = 0\) if \(|~\bigcup _{i=1}^{k}{L(G_i)}~| = 1\) (Stop condition);
-
2.
Otherwise, \( MinSGT(G_1,\ldots ,G_k) =\)
Note that, given a bipartition \((B_l,B_r) \in \mathscr {B}(G_1,\ldots ,G_k)\), for each i such that \(1 \le i \le k\), \(G_{i|B_l}\) and \(G_{i|B_r}\) are equal either to \(\emptyset \) or \(G_i\) or \(G_{i_l}\) or \(G_{i_r}\). Thus, \(G_{i|B_l}\) and \(G_{i|B_r}\) are always either empty trees or complete subtrees of \(G_i\). Furthermore, the existence of a compatible bipartition, at each step, follows from the fact that the input gene trees are assumed to be consistent.
In [41] we show how Theorem 2 can be modified to account for inconsistencies between gene trees, by adding a third equation: If \(|~\bigcup _{i=1}^{k}{L(G_i)}~| > 1\) and \(|~\mathscr {B}(G_1,\ldots ,G_k)~| = 0\), \(MinSGT(G_1,\ldots ,G_k) = +\infty \). We also show that \(|\mathscr {B}(G_1,\ldots ,G_k)| \le (\frac{4^k}{2})-1\), resulting in the time complexity of the overall algorithm being \(O((n+1)^k \times 4^k \times k)\), where n is the maximum number of nodes in a tree \(G_i\).
5.7 A Unifying View for the DL Model
The polytomy-based and supertree-based framework for gene tree correction have been developed separately, considering separate assumptions and constraints. In the absence of a unifying model, the conservative or permissive nature of each framework with respect to the other can only be tested empirically. A conceptual breakthrough is the discovery that, for the DL model, the two frameworks are in fact two special cases of a more general one: LabelGTC expressed in terms of a 0–1 edge-labeled gene tree [26], and TripletGTC expressed in terms of preserving triplets [26]. Here, we focus on LabelGTC.
Given an initial tree G for a gene family \({\varGamma }\), the correction problem can be defined as finding a “better tree” \(G'\) according to a reconciliation cost. The various versions of the problem differ on the flexibility we have in modifying G. Regarding which parts of G should be preserved, an intuitive way is to take advantage of the support on each branch (x, y) which reflects the confidence we have on \(L(G_y)\) being a separate clade in the gene family. Hence, we could allow modifications only on weakly supported branches, i.e the ones with a support below a given threshold, while preserving all well-supported branches. Using a threshold, we therefore obtain a 0–1 edge-labeling of E(G), where 0 indicates a low support and 1 a high support.
If G further contains a set of separated subtrees whose topologies are to be “trusted”, they should also be preserved during correction. For example, ortholog groups that agree with the species tree and were separately obtained to build G may be trusted.
Accordingly, we describe below the most general gene tree correction problem (see Fig. 5.8 for an illustration), where a covering set of subtrees \({\mathscr {C}_G}\) for G is a set of separated subtrees of G, \({\mathscr {C}_G}=\{G_{x_1},G_{x_2},\ldots ,G_{x_n}\}\) such that \(\bigcup _{i=1}^{n}L(G_{x_i})= L(G)\), and a 0–1 edge-labeling for G is a function f defined from the set of edges E(G) to \(\{0,1\}\). In the following formulation, edge labels are ignored for the trees of \({\mathscr {C}_G}\). For an extension that considers edge-labeling inside the covering set, see [26].
Label Respecting Gene Tree Correction (LabelGTC) Problem:
Input: A species tree S, a gene tree G, a covering set of trees \({\mathscr {C}_G}\) for G and a 0–1 edge-labeling f for G.
Output: A supertree \(G'\) for \({\mathscr {C}_G}\) of minimum reconciliation cost such that: if \((x,y) \in E(G)\setminus E({\mathscr {C}_G})\) and \(f(x,y)=1\), then there is an edge \((x',y')\) in \(E(G')\) such that \(L(G_y) = L(G'_{y'})\).
When no information on “trusted” separated subtrees is available, each tree of \({\mathscr {C}_G}\) is simply restricted to a leaf of G, and \({\mathscr {C}_G}\) thus refers to the leafset of G.
(Figure modified from [26]; use permitted under the Creative Commons Attribution License CC-BY 3.0) Left. A species tree S for \(\varSigma = \{a,b,c,d,e\}\), a reconciled 0–1 edge-labeled gene tree G for \({\varGamma }= \{a_1,b_1,c_1,c_2,d_1,d_2,d_3,e_1,e_2,e_3\}\) where each leaf \(x_i\) denotes a gene belonging to genome x, and a covering set \({\mathscr {C}_G}\) of subtrees for G indicated by blue circles around each subtree. Rectangular nodes represent duplications, black dots are speciations and dotted lines are losses. Right. A supertree for \({\mathscr {C}_G}\) of minimum reconciliation cost (cost of 3) respecting the edge-labeling of G
In the following, we reformulate the polytomy-related (Sect. 5.5) and supertree-related (Sect. 5.6.2) correction problems according to a 0–1 edge-labeled gene tree (see Fig. 5.9 for an illustration of the problems). We then show that they are special cases of the general LabelGTC problem.
(Figure from [26]; use permitted under the Creative Commons Attribution License CC-BY 3.0) A species tree S for \(\varSigma = \{a,b,c,d,e\}\) and a gene tree G for \({\varGamma }= \{a_1,b_1,c_1,c_2,d_1,d_2,d_3,e_1,e_2,e_3\}\) with a covering set \({\mathscr {C}_G}\) of subtrees for G as in Fig. 5.8 (without the 0–1 labeling of edges). Bottom left. A polytomy resolution for \({\mathscr {C}_G}\) of minimum reconciliation cost (cost of 3). Bottom right. A supertree for \({\mathscr {C}_G}\) of minimum reconciliation cost (cost of 2). Top right. A triplet-respecting supertree for \({\mathscr {C}_G}\) of minimum reconciliation cost (cost of 5). Note that the solutions for the TRS, SGT and PolyRes problems may differ from the optimal supertree for the LabelGTC problem, because of the 0–1 edge-labeling. In this particular case, the optimal supertree for the SGT problem is identical to the one returned for LabelGTC in Fig. 5.8
In the general version of the polytomy resolution problem, all weakly supported internal branches of G are contracted, leading to a non-binary tree \(G^{nb}\). The goal is then to find a binary refinement of \(G^{nb}\) minimizing the reconciliation cost.
Mutiple Polytomy Resolution (M-PolyRes) Problem:
Input: A species tree S and a 0–1 edge-labeled gene tree G and the tree \(G^{nb}\) obtained from G by contracting edges labeled 0;
Output: A binary refinement of \(G^{nb}\) minimizing the reconciliation cost.
In the simplest form of the polytomy resolution problem, we have a single polytomy which consists of a non-binary node at the root of \(G^{nb}\). The subtrees rooted at the children of \(r(G^{nb})\) are the “trusted” partial trees that should remain subtrees of the final tree (see the tree obtained from PolyRes in Fig. 5.9).
Polytomy Resolution (PolyRes) Problem:
Input: A species tree S, a gene tree G and a covering set of trees \({\mathscr {C}_G}\) for G.
Output: A supertree \(G'\) for \({\mathscr {C}_G}\) of minimum reconciliation cost such that for any tree \(G_i \in {\mathscr {C}_G}\), \(G'_{|L(G_i)}= G_i\).
Now recall the MinSGT correction problem introduced in Sect. 5.6.2, but in the simplest case of separated gene trees.
SuperGenetree (SGT) Problem:
Input: A species tree S, a gene tree G and a covering set of trees \({\mathscr {C}_G}\) for G.
Output: A supertree \(G'\) for \({\mathscr {C}_G}\) of minimum reconciliation cost.
To avoid having a supertree grouping genes that are far apart in the original tree, we also introduced, in [41], an alternative version of the problem restricting the output space to supertrees preserving the topology of any triplet of genes taken from three different input subtrees of \({\mathscr {C}_G}\). A formulation of the triplet-based constrained supertree problem follows.
Triplet-Respecting SuperGeneTree (TRS) Problem:
Input: A species tree S, a gene tree G and a covering set of trees \({\mathscr {C}_G}\) for G.
Output: A supertree \(G'\) for \({\mathscr {C}_G}\) of minimum reconciliation cost respecting the following property: for any triplet (a, b, c) where a, b and c are genes of \({\varGamma }\) being leaves of three different trees of \({\mathscr {C}_G}\), \(G'_{|\{a,b,c\}}= G_{|\{a,b,c\}}\).
The difference between the TRS and SGT problems is illustrated in Fig. 5.9. The solution of the SGT Problem shown in that figure is not a solution of the TRS problem as the triplet \((a_1, c_1,c_2)\), where each gene belongs to a separate subtree of \({\mathscr {C}_G}\), has the topology \((a_1, (c_1,c_2))\) in the SGT tree while it has the topology \(((a_1,c_1),c_2)\) in G.
(Figure from [26]; use permitted under the Creative Commons Attribution License CC-BY 3.0) (1) A gene tree G with a covering set \({\mathscr {C}_G}\) composed of 7 subtrees indicated as triangles. The set \(E(G)\setminus E({\mathscr {C}_G})\) contains 7 terminal edges (dotted lines) and 5 non-terminal edges (solid lines). (2), (3) and (4) are three 0–1 edge-labeling corresponding respectively to the PolyRes, SGT and TRS problems. (5) is a general input of the LabelGTC problem
A unifying view: Theorem 3 shows that the polytomy-related and supertree-related problems are in fact special cases of the general LabelGTC problem. We begin by introducing some notation.
Given a covering set of subtrees \({\mathscr {C}_G}\) for G, we say that an edge (x, y) of \(E(G)\setminus E({\mathscr {C}_G})\) is a terminal edge if y is the root of a tree in \({\mathscr {C}_G}\). Any other edge in \(E(G)\setminus E({\mathscr {C}_G})\) is called a non-terminal edge (see Fig. 5.10 for an illustration).
Theorem 3
Let G be a 0–1 edge-labeled gene tree and \({\mathscr {C}_G}\) be a covering set for G. Then the LabelGTC Problem is reduced to the:
-
1.
M-PolyRes Problem if \({\mathscr {C}_G}= L(G)\); Otherwise:
-
2.
PolyRes Problem if all non-terminal edges are labeled 0, and all terminal edges are labeled 1;
-
3.
SGT Problem if all non-terminal and terminal edges are labeled 0;
-
4.
TRS Problem if all non-terminal edges are labeled 1, and all terminal edges are labeled 0.
Finally, we have developed an algorithm, called LabelGTC, handling the general version of the problem, not represented by any of the special cases reflected in Theorem 3. For any edge (x, y) in \(E(G)\setminus E({\mathscr {C}_G})\) labeled 1, there should exist a node \(y'\) in the final corrected tree \(G'\) such that \(L(y')\) = L(y). So the subtree \(G'_{y'}\) of \(G'\) for the subset \(L(G_y)\) can first be constructed independently of the remaining nodes of \(G'\), and then grafted at the appropriate location in a way minimizing the reconciliation cost. The LabelGTC algorithm proceeds iteratively, in a bottom-up order, on subtrees \(G_{y}\) with parental edge (x, y) fitting the above criterion, and is recursively called to reconstruct \(G'_{y'}\). Each solution \(G'_{y'}\) is implicitly treated as a leaf in subsequent calls to avoid modifying its content.
In [26], we showed that the time complexity of the algorithm is related to the time complexity of MinSGT, which makes it exponential in the number of terminal subtrees. More precisely, the algorithm runs in time \(O(4^k \cdot (n+1)^k \cdot k)\), where \(n = |{\varGamma }|\) and \(k = |{\mathscr {C}_G}|\).
5.8 Discussion
Efficient pipelines for gene tree inference should typically include accurate gene sequence alignment tools and use inference methods combining information from both micro-evolutionary (sequence level) and macro-evolutionary (genome level) information. In the recent years, new algorithms improving the accuracy of sequence alignment and gene tree inference have been described.
In particular, probabilistic gene tree construction methods relying on complex evolutionary models that account for both sequence and species tree data have been developed [2, 4, 66, 67, 76, 84, 86]. These methods unfortunately present some drawbacks inherent to probabilistic methods, namely the huge computational time associated with the numerical integration of the likelihood, and the prior analyses required to satisfy the input requirements (e.g., dating the species tree).
In practice, alternative parsimony-based approaches, a posteriori correcting gene trees inferred from sequence-only data with species tree information, are used instead. Such algorithms, although limited in some aspects when compared to probabilistic ones, have consistently produced trees with high accuracy, while being much faster. This time efficiency allows applying the correction method to a wide set of data. For example, in [60], we used ProfileNJ to correct the PhyML trees built on the whole Ensembl Compara gene families (20,519 families in total). According to several criteria, including likelihood, reconciliation score, and ancestral genome content, these corrected trees constitute an arguably better dataset than the one stored in the Ensembl database.
Another advantage of parsimony methods is that they can be easily extended to consider other sources of information. For example, gene order may provide information on gene orthology and paralogy. In fact, two synteny blocks, i.e., two chromosomal segments (in the same genome or in two different genomes) containing genes form the same gene families are likely to have a common ancestor. Depending on whether they diverged from a speciation or a duplication event, gene pairs in the two synteny blocks will either be all orthologs or all paralogs. This information has been considered for correcting a gene tree in [44, 46].
Alternatively, functional similarity between genes is also, usually, a good indicator for orthology [3, 29]. We are presently exploring ways to efficiently use scores based on Gene Ontology annotations to establish terminal preserved trees in LabelGTC.
The main difficulty remains how to integrate all the developed algorithmic tools, each handling a given type of information on genes and trees, into a single robust framework for gene tree reconstruction. In addition, rather than applying corrections in an incremental manner, with the risk of obtaining very different trees depending on the order of execution, the challenge is to consider the variety of sequence, functional, order and evolutionary information all together in a single algorithm. The LabelGTC algorithm, considering polytomy resolution and supertree reconstruction in a unifying framework is an effort in this direction. However, fitness to sequence information may still be lost after correction, unless we constraint the output to be statistically equivalent to the best maximum likelihood tree.
Therefore, approaches suitable for the resolution of Multi-Objective Optimization Problems (MOOP) have to be explored. In this context, we have developed GATC [59], a genetic algorithm minimizing a measure combining both tree likelihood (according to sequence evolution) and a reconciliation score that accounts for HGT. An advantage of this approach is its ability to improve search efficiency by exploring a population of trees at each step. Although much slower than deterministic methods for correction, GATC outperforms all these methods in terms of accuracy.
(i) The same evolution with ILS represented in Fig. 5.4; (ii) The locus tree inferred by DLCpar [95], inducing one duplication at the root and one loss. (iii) An alternative explanation of the gene tree with ILS, the duplication occurring lower in the species tree, and no loss. This most parsimonious DL history with ILS is not inferred by DLCpar, however the hill-climbing heuristic described in the same paper did find it.; (iv) The gene tree/species tree reconciliation leading to two duplications and four losses; (v) The set of largest speciation subtrees in the gene tree; (v) The tree obtained by MinSGT reflecting the most parsimonious history represented in (iii)
From an algorithmic point of view, a lot remains to be done. Unifying the diversity of evolutionary models and datasets is still far from being reached and raises the interesting problem of how we can simultaneously account, in the same evolutionary model, for sequence evolution as well as duplications, losses, HGTs, recombination, hybridization, and ILS. Interestingly, some of the methods developed for these events, often taken separately, might be more related than expected. For example, as we show in Fig. 5.11, the parsimony method described in [95] for reconciling a binary gene tree with a binary species tree, while accounting for duplications, losses and ILS (see Sect. 5.3.3), may be compared to the strategy using MinSGT that we explored in Sect. 5.6.2. The latter consists of removing upper branches of the gene tree, keeping speciation trees, i.e., subtrees with only speciation nodes, and then using a supertree method to reconstruct a most parsimonious supertree containing them all. To which extent the two methods are comparable from a theoretical point of view? How can the supertree method be applied to account for ILS? Can we take advantage of the similarity between the two problems to design more efficient algorithms than the exponential dynamic programming algorithm developed for MinSGT? These are few questions that will be considered in future developments.
As we have no direct access to the past, it is difficult to objectively evaluate the accuracy of gene tree reconstruction methods. The most intuitive way is to compare inferences on simulated gene families, where the “true” evolutionary histories according to some given model of evolution with controlled parameters, are known. Aside from tree topology comparison using metrics such as the Robinson-Foulds distance [63, 68], we can also assess how close the evolutionary scenarios inferred are to the true ones. In [60], we have additionally considered metrics based on ancestral gene content inferred from reconciliation, and ancestral gene adjacencies [9]. The latter is particularly useful as measure for gene tree accuracy for linear genomes, given that at most two adjacencies per gene copy should be expected.
Since good results on simulated datasets do not guarantee the same on real ones, as they may not conform to the evolutionary model used for simulations, well-studied gene families for which good trees are available have been used to construct reference datasets. In this regard, several ongoing works, such as the SwissTree [12] project, are undertaking great efforts to provide manually curated “gold standard” gene trees. However, the number of available “gold standard” remains extremely low (19 in SwissTree) and does not allow extensive covering of the many and intricate pathways of gene evolution. Therefore, developing new sophisticated frameworks, accounting for various gene characteristics for producing good benchmarks, is still needed.
Notes
- 1.
See Quest for Orthologs links at http://questfororthologs.org/.
References
Aho, A., Yehoshua, S., Szymanski, T., Ullman, J.: Inferring a tree from lowest common ancestors with an application to the optimization of relational expressions. SIAM J. Comput. 10(3), 405–421 (1981)
Akerborg, O., Sennblad, B., Arvestad, L., Lagergren, J.: Simultaneous bayesian gene tree reconstruction and reconciliation analysis. Proc. Nal. Acad. Sci. USA 106(14), 5714–5719 (2009)
Altenhoff, A.M., Studer, R.A., Robinson-Rechavi, M., Dessimoz, C.: Resolving the ortholog conjecture: orthologs tend to be weakly, but significantly, more similar in function than paralogs. PLoS Comput. Biol. 8(5), e1002,514 (2012)
Arvestad, L., Berglund, A., Lagergren, J., Sennblad, B.: Gene tree reconstruction and orthology analysis based on an integrated model for duplications and sequence evolution. In: RECOMB, pp. 326–335 (2004)
Bader, D., Moret, B., Yan, M.: A linear-time algorithm for computing inversion distance between signed permutations with an experimental study. J. Comput. Biol. 8(5), 483–491 (2001)
Bansal, M., Alm, E., Kellis, M.: Efficient algorithms for the reconciliation problem with gene duplication, horizontal transfer and loss. Bioinformatics 28(12), i283–i291 (2012). https://doi.org/10.1093/bioinformatics/bts225
Bansal, M., Burleigh, J., Eulenstein, O., Fernández-Baca, D.: Robinson-foulds supertrees. Alg. Mol. Biol. 5(18) (2010)
Bansal, M., Wu, Y., Alm, E., Kellis., M.: Improved gene tree error-correction in the presence of horizontal gene transfer. Bioinformatics 31(8), 1211–1218 (2015). https://doi.org/10.1093/bioinformatics/btu806
Bérard, S., Gallien, C., Boussau, B., Szollosi, G., Daubin, V., Tannier, E.: Evolution of gene neighborhoods within reconciled phylogenies. Bioinformatics 28(18), i382–i388 (2012)
Berglund, A., Sjolund, E., Ostlund, G., Sonnhammer, E.: InParanoid 6: eukaryotic ortholog clusters with inparalogs. Nucl. Acid Res. 36 (2008)
Bininda-Emonds, O. (ed.): Phylogenetic Supertrees combining information to reveal The Tree of Life. In: Computational Biology. Kluwer Academic, Dordrecht, The Netherlands (2004)
Boeckmann, B., Robinson-Rechavi, M., Xenarios, I., Dessimoz, C.: Conceptual framework and pilot study to benchmark phylogenomic databases based on reference gene trees. Brief. Bioinform. 12(5), 423–435 (2011)
Bork, D., Cheng, R., Wang, J., Sung, J., Libeskind-Hadas, R.: On the computational complexity of the maximum parsimony reconciliation problem in the duplication-loss-coalescence model. Algorithms Mol. Biol. 12(1), 6 (2017)
Boussau, B., Szöllősi, G., Duret, L., Gouy, M., Tannier, E., Daubin, V.: Genome-scale coestimation of species and gene trees. Genome Res. 23, 323–330 (2013)
Chan, Y., Ranwez, V., Scornavacca, C.: Exploring the space of gene/species reconciliations with transfers. J. Math. Biol. 71(5), 1179–1209 (2015)
Chan, Y., Ranwez, V., Scornavacca, C.: Inferring incomplete lineage sorting, duplications, transfers and losses with reconciliations. J. Theoret. Biol. 432, 1–13 (2017)
Chang, W., Eulenstein, O.: Reconciling gene trees with apparent polytomies. In: Chen, D., Lee, D.T. (eds.) Proceedings of the 12th Conference on Computing and Combinatorics (COCOON). Lecture Notes in Computer Science, vol. 4112, pp. 235–244 (2006)
Chen, K., Durand, D., Farach-Colton, M.: Notung: dating gene duplications using gene family trees. J. Comput. Biol. 7, 429–447 (2000)
Constantinescu, M., Sankoff, D.: An efficient algorithm for supertrees. J. Classif. 12, 101–112 (1995)
Darby, C.A., Stolzer, M., Ropp, P.J., Barker, D., Durand, D.: Xenolog classification. Bioinformatics 33(5), 640–649 (2016)
David, L., Alm, E.: Rapid evolutionary innovation during an Archaean genetic expansion. Nature 469 (2011)
Doyon, J.P., Chauve, C., Hamel, S.: Space of gene/species trees reconciliations and parsimonious models. J. Comput. Biol 16(10), 1399–1418 (2009)
Doyon, J., Ranwez, V., Daubin, V., Berry, V.: Models, algorithms and programs for phylogeny reconciliation. Brief. Bioinform. 12(5), 392–400 (2011)
Doyon, J.P., Scornavacca, C., Gorbunov, K.Y., Szöllősi, G.J., Ranwez, V., Berry, V.: An efficient algorithm for gene/species trees parsimonious reconciliation with losses, duplications and transfers. In: Tannier, E. (ed.) RECOMB International Workshop on Comparative Genomics, RECOMB-CG, pp. 93–108. Springer (2010)
Durand, D., Halldórsson, B.V., Vernot, B.: A hybrid micro-macroevolutionary approach to gene tree reconstruction. J. Comput. Biol. 13(2), 320–335 (2006)
El-Mabrouk, N., Ouangraoua, A.: A general framework for gene tree correction based on duplication-loss reconciliation. In: LIPIcs, Workshop on Algorithms in Bioinformatics (WABI), vol. 88, pp. 8:1–8:14 (2017)
Fitch, W.: Homology—a personal view on some of the problems. Trends Genet. 16(5), 227–231 (2000)
Flicek, P., et al.: Ensembl 2012. Nucleic Acids Res. 40, D84–D90 (2012)
Gabaldón, T., Koonin, E.V.: Functional and evolutionary implications of gene orthology. Nat. Rev. Genet. 14(5), 360 (2013)
Goodman, M., Czelusniak, J., Moore, G., Romero-Herrera, A., Matsuda, G.: Fitting the gene lineage into its species lineage, a parsimony strategy illustrated by cladograms constructed from globin sequences. Syst. Zool. 28, 132–163 (1979)
Górecki, P., Eulenstein, O.: Algorithms: simultaneous error-correction and rooting for gene tree reconciliation and the gene duplication problem. BMC Bioinform. 13(Supp 10), S14 (2011)
Gorecki, P., Eulenstein, O., Tiuryn, J.: Unrooted tree reconciliation: a unified approach. IEEE/ACM Trans. Comput. Biol. Bioinform. 10(2), 522–536 (2013)
Guindon, S., Gascuel, O.: A simple, fast and accurate algorithm to estimate large phylogenies by maximum likelihood. Syst. Biol. 52, 696–704 (2003)
Hallett, M., Lagergren, J.: Efficient algorithms for lateral gene transfer problems. In: Proceedings of the Fifth Annual International Conference on Computational Biology, RECOMB-CG, pp. 149–156 (2001)
Höhna, S., Drummond, A.J.: Guided tree topology proposals for bayesian phylogenetic inference. Syst. Biol. 61(1), 1–11 (2011)
Jacox, E., Chauve, C., Szöllősi, G.J., Ponty, Y., Scornavacca, C.: ecceTERA: comprehensive gene tree-species tree reconciliation using parsimony. Bioinformatics 32(13), 2056–2058 (2016). https://doi.org/10.1093/bioinformatics/btw105
Jacox, E., Weller, M., Tannier, E., Scornavacca, C.: Resolution and reconciliation of non-binary gene trees with transfers, duplications and losses. Bioinformatics 33(7), 980–987 (2017)
Kordi, M., Bansal, M.: On the complexity of duplication-transfer-loss reconciliation with non-binary gene trees. IEEE/ACM Trans. Comput. Biol. Bioinform. (2016)
Kordi, M., Bansal, M.: Exact algorithms for duplication-transfer-loss reconciliation with non-binary gene trees. IEEE/ACM Trans. Comput. Biol. Bioinform. (2017)
Lafond, M., Chauve, C., Dondi, R., Manuel, El-Mabrouk, N.: Polytomy refinement for the correction of dubious duplications in gene trees. Bioinformatics 30(17), i519–i526 (2014)
Lafond, M., Chauve, C., El-Mabrouk, N., Ouangraoua, A.: Gene tree construction and correction using supertree and reconciliation. IEEE/ACM Trans. Comput. Biol. Bioinform. (TCBB) PP(99), 12 pp. (2018)
Lafond, M., Noutahi, E., El-Mabrouk, N.: Efficient non-binary gene tree resolution with weighted reconciliation cos. In: 27th Annual Symposium on Combinatorial Pattern Matching (CPM) (2016)
Lafond, M., Ouangraoua, A., El-Mabrouk, N.: Reconstructing a supergenetree minimizing reconciliation. BMC Genomics 16, S4 (2015). Special issue of RECOMB-CG 2015
Lafond, M., Semeria, M., Swenson, K., Tannier, E., El-Mabrouk, N.: Gene tree correction guided by orthology. BMC Bioinform. 14(supp 15)(S5) (2013)
Lafond, M., Swenson, K., El-Mabrouk, N.: An optimal reconciliation algorithm for gene trees with polytomies. In: WABI. LNCS, vol. 7534, pp. 106–122 (2012)
Lafond, M., Swenson, K., El-Mabrouk, N.: Error detection and correction of gene trees. In: Models and Algorithms for Genome Evolution. Springer (2013)
Lai, H., Stolzer, M., Durand, D.: Fast heuristics for resolving weakly supported branches using duplication, transfers, and losses. In: RECOMB-CG, 22 pp. (2017)
Lartillot, N., Philippe, H.: A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21(6), 1095–1109 (2004). http://dx.doi.org/10.1093/molbev/msh112
Lechner, M., Findeiß, S., Steiner, L., Manja, M., Stadler, P., Prohaska, S.: Proteinortho: Detection of co-orthologs in large-scale analysis. BMC Bioinform. 12(1), 1 (2011)
Li, L., Stoeckert, C.J., Roos, D.: OrthoMCL: identification of ortholog groups for eukaryotic genomes. Genome Res. 13, 2178–2189 (2003)
Libeskind-Hadas, R., Charleston, M.: On the computational complexity of the reticulate cophylogeny reconstruction problem. J. Comput. Biol. 16 (2009)
Maddison, W.P.: Gene trees in species trees. Syst. Biol. 46(3), 523–536 (1997)
Massey, S., Churbanov, A., Rastogi, S., Liberles, D.: Characterizing positive and negative selection and their phylogenetic effects. Gene 418, 22–26 (2008)
Moret, B., Warnow, T.: Molecular evolution: producing the biochemical data. In: Zimmer, E., Roalson, E. (eds.) Methods in Enzymology, Part B, vol. 395, pp. 673–700. Elsevier (2005)
Moret, B.M., Bader, D.A., Wyman, S., Warnow, T., Yan, M.: A new implementation and detailed study of breakpoint analysis. In: Biocomputing 2001, pp. 583–594. World Scientific (2000)
Ng, M., Wormald, N.: Reconstruction of rooted trees from subtrees. Discrete Appl. Math. 69, 19–31 (1996)
Nguyen, N., Mirarab, S., Warnow, T.: MRL and SuperFine+MRL: new supertree methods. Algorithms Mol. Biol. 7(3) (2012)
Nguyen, T.H., Ranwez, V., Pointet, S., Chifolleau, A.M.A., Doyon, J.P., Berry, V.: Reconciliation and local gene tree rearrangement can be of mutual profit. Algorithms Mol. Biol. 8(1), 12 (2013). http://dx.doi.org/10.1186/1748-7188-8-12
Noutahi, E., El-Mabrouk, N.: GATC: a genetic algorithm for gene tree construction under the duplication-transfer-loss model of evolution. BMC Genomics 19(2), 102 (2018)
Noutahi, E., Semeria, M., Lafond, M., Seguin, J., Gueguen, L., El-Mabrouk, N., Tannier, E.: Efficient gene tree correction guided by genome evolution. PLoS One 11(8) (2016)
Ovadia, Y., Fielder, D., Conow, C., Libeskind-Hadas, R.: The cophylogeny reconstruction problem is NP-complete. J. Comput. Biol. 18(1), 59–65 (2011). https://doi.org/10.1089/cmb.2009.0240
Page, R.D., Cotton, J.A.: Genetree: a tool for exploring gene family evolution. In: Comparative Genomics, pp. 525–536. Springer (2000)
Pattengale, N., Gottlieb, E., Moret, B.: Efficiently computing the Robinson-Foulds metric. J. Comput. Biol. 14(6), 724–735 (2007)
Ranwez, V., Berry, V., Criscuolo, A., Fabre, P., Guillemot, S., Scornavacca, C., Douzery, E.: PhySIC: a veto supertree method with desirable properties. Syst. Biol. 56(5), 798–817 (2007)
Ranwez, V., Criscuolo, A., Douzery, E.: SuperTriplets: a triplet-based supertree approach to phylogenomics. Bioinformatics 26(12), i115–i123 (2010)
Rasmussen, M., Kellis, M.: A Bayesian approach for fast and accurate gene tree reconstruction. Mol. Biol. Evol. 28(1), 273–290 (2010)
Rasmussen, M.D., Kellis, M.: Unified modeling of gene duplication, loss, and coalescence using a locus tree. Genome Res. 22(4), 755–765 (2012)
Robinson, D., Foulds, L.: Comparison of phylogenetic trees. Math. Biosci. 53, 131–147 (1981)
Rodrìguez-Ezpeleta, N., Brinkmann, H., Roure, B., Lartillot, N., Lang, B.F., Philippe, H.: Detecting and overcoming systematic errors in genome-scale phylogenies. Syst. Biol. 56(3), 389–399 (2007). http://dx.doi.org/10.1080/10635150701397643
Rogers, J., Fishberg, A., Youngs, N., Wu, Y.C.: Reconciliation feasibility in the presence of gene duplication, loss, and coalescence with multiple individuals per species. BMC Bioinform. 18(1), 292 (2017)
Ronquist, F., Huelsenbeck, J.: MrBayes3: Bayesian phylogenetic inference under mixed models. Bioinformatics 19, 1572–1574 (2003)
Roshan, U., Moret, B., Warnow, T., Williams, T.: Performance of supertree methods on various dataset decompositions. In: Bininda-Edmonds, O. (ed.) Phylogenetic Supertrees: Combining Information to Reveal the Tree of Life, pp. 301–328. Springer (2004)
Scornavacca, C., van Iersel, L., Kelk, S., Bryant, D.: The agreement problem for unrooted phylogenetic trees is FPT. J. Graph Algorithms Appl. 18(3), 385–392 (2014)
Scornavacca, C., Jacox, E., Szollosi, G.: Joint amalgamation of most parsimonious reconciled gene trees. Bioinformatics 31(6), 841–848 (2015)
Semple, C.: Reconstructing minimal rooted trees. Discrete Appl. Math. 127(3) (2003)
Sjöstrand, J., Tofigh, A., Daubin, V., Arvestad, L., Sennblad, B., Lagergren, J.: A Bayesian method for analyzing lateral gene transfer. Sys. Biol. 63(3), 409–420 (2014)
Skovgaard, M., Kodra, J., Gram, D., Knudsen, S., Madsen, D., Liberles, D.: Using evolutionary information and ancestral sequences to understand the sequence-function relationship in GLP-1 agonists. J. Mol. Biol. 363, 977–988 (2006)
Stamatakis, A.: RAxML-VI-HPC: maximum likelihood-based phylogenetic analysis with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006)
Steel, M.: The complexity of reconstructing trees from qualitative characters and subtrees. J. Classif. 9, 91–116 (1992)
Steel, M., Rodrigo, A.: Maximum likelihood supertrees. Syst. Biol. 57(2), 243–250 (2008)
Stolzer, M., Lai, H., Xu, M., Sathaye, D., Vernot, B., Durand, D.: Inferring duplications, losses, transfers and incomplete lineage sorting with nonbinary species trees. Bioinformatics 28(18), i409–i415 (2012)
Swenson, K.M., El-Mabrouk, N.: Gene trees and species trees: irreconcilable differences. BMC Bioinform. 13(Suppl 19), S15 (2012)
Swenson, M., Suri, R., Linder, C., Warnow, T.: SuperFine: fast and accurate supertree estimation. Sys. Biol. 61(2), 214–227 (2012). Special issue RECOMB-CG 2012
Szöllősi, G., Rosikiewicz, W., Boussau, B., Tannier, E., Daubin, V.: Efficient exploration of the space of reconciled gene trees. Syst. Biol. 62(6), 901–912 (2013). http://dx.doi.org/10.1093/sysbio/syt054
Szöllősi, G., E., Tannier, Daubin, V., Boussau, B.: The inference of gene trees with species trees. Syst. Biol. 64(1), e42–e62 (2014)
Szöllősi, G.J., Tannier, E., Lartillot, N., Daubin, V.: Lateral gene transfer from the dead. Syst. Biol. 62(3), 386–397 (2013)
Tatusov, R., Galperin, M., Natale, D., Koonin, E.: The COG database: a tool for genome-scale analysis of protein functions. Nucleic Acids Res. 28, 33–36 (2000)
Taylor, S., de la Cruz, K., Porter, M., Whiting, M.: Characterization of the long-wavelength opsin from Mecoptera and Siphonaptera: does a flea see? Mol. Biol. Evol. 22, 1165–1174 (2005)
Thomas, P.: GIGA: a simple, efficient algorithm for gene tree inference in the genomic age. BMC Bioinform. 11, 312 (2010)
Tofigh, A.: Using trees to capture reticulate evolution: lateral gene transfers and cancer progression. Ph.D. thesis, KTH Royal Institute of Technology, Sweden (2009)
Tofigh, A., Hallett, M., Lagergren, J.: Simultaneous identification of duplications and lateral gene transfers. IEEE/ACM Trans. Comput. Biol. Bioinform. 8(2), 517–535 (2011). https://doi.org/10.1109/TCBB.2010.14
Vernot, B., Stolzer, M., Goldman, A., Durand, D.: Reconciliation with non-binary species trees. J. Comput. Biol. 15, 981–1006 (2009)
Wu, T., Zhang, L.: Structural properties of the reconciliation space and their applications in enumerating nearly-optimal reconciliations between a gene tree and a species tree. BMC Bioinform. 12, S7 (2011)
Wu, Y., Rasmussen, M., Bansal, M., Kellis, M.: TreeFix: statistically informed gene tree error correction using species trees. Syst. Biol. 62(1), 110–120 (2013)
Wu, Y., Rasmussen, M., Bansal, M., Kellis, M.: Most parsimonious reconciliation in the presence of gene duplication, loss, and deep coalescence using labeled coalescent trees. Genome Res. 24, 475–486 (2014)
Zhang, L.: On Mirkin-Muchnik-Smith conjecture for comparing molecular phylogenies. J. Comput. Biol. 4, 177–188 (1997)
Zheng, Y., Wu, T., Zhang, L.: Reconciliation of gene and species trees with polytomies (2012). arXiv:1201.3995
Zheng, Y., Zhang, L.: Reconciliation with non-binary gene trees revisited. In: Proceedings of RECOMB. Lecture Notes in Computer Science, vol. 8394, pp. 418–432 (2014)
Zmasek, C.M., Eddy, S.R.: A simple algorithm to infer gene duplication and speciiation events on a gene tree. Bioinformatics 17, 821–828 (2001)
Acknowledgements
The authors acknowledge the support of the Fonds de Recherche du Québec Nature et Technologie (FRQNT) and of the Natural Sciences and Engineering Research Council (NSERC) (Discovery Grant RGPIN-249834).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this chapter
Cite this chapter
El-Mabrouk, N., Noutahi, E. (2019). Gene Family Evolution—An Algorithmic Framework. In: Warnow, T. (eds) Bioinformatics and Phylogenetics. Computational Biology, vol 29. Springer, Cham. https://doi.org/10.1007/978-3-030-10837-3_5
Download citation
DOI: https://doi.org/10.1007/978-3-030-10837-3_5
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-10836-6
Online ISBN: 978-3-030-10837-3
eBook Packages: Computer ScienceComputer Science (R0)