Abstract
Lung cancer ranks as second most prevalent type of cancer. Still predictions for survival of lung cancer patients are not accurate. In this research, we try to create a prediction model, with the help of machine learning to accurately predict the survival of non-small cell lung cancer patients (NSCLC). Clinical data of 559 patients was taken for training and testing of models. We have developed multilevel perceptron model for survival prediction. Other models developed during this study were compared to measure performance of our model. Attributes that are found to be useful as biomarkers for prediction of survival analysis of NSCLC have also been computed and ranked accordingly for increase in accuracy of prediction model by implementing feature selection method. The final model included T stage, N stage, Modality, World Health Organization Performance status, Cumulative Total Tumor dose, tumor load, Overall treatment time as the variables. Two year survival was chosen as the prediction outcome. Neural Network was found as the best prediction model with area under Curve (AUC) of 0.75. By far to our knowledge Multilevel Neural Network is found to be the best model for predicting two-year survival among patients of non-small cell lung cancer.
Access provided by Autonomous University of Puebla. Download conference paper PDF
Similar content being viewed by others
Keywords
- Multilevel neural network
- Non-small cell lung cancer
- Machine learning
- Feature selection
- ReliefF
- Survival prediction
1 Introduction
Lung cancer is second most prevalent type of cancer. With 2,34,030 new cases being recorded alone in United States as per 2018 [1]. Non-small cell lung cancer shares 80–85% from the reported cases [1]. One of the critical concerns for a patient diagnosed with cancer is the survival expectancy [2]. At present 5 year survival expectation of lung cancer patients is 15%, stage I has 5 year survival expectation of 70%, 45% in stage II and 10–30% for stage III, while that for IIIB and IV are respectively 10% and < 5% [1].
Though there have been notable advancements in technology still survival prediction uses basic methods. Currently TNM staging is used for survival prediction, although its primary use was to suggest operability scheme that can be performed on patient [3]. Where T stage describes the extent and size of main (primary) tumor, N stage refers to nearby (regional) lymph nodes that have cancer, M stage refers to whether the cancer has metastasized. Other predictive models such as Naïve Bayes model, Random Forest, Support Vector Machine have been proposed previously for survival prediction [3, 4] still a desirable accuracy is not achieved. Feature selection in previously developed models such as, Naïve Bayes model was based on expert opinion [3], while Random Forest model implemented feature selection by using ReliefF algorithm [4].
This study aims to improve the 2-year survival prediction for patients suffering from NSCLC. Features which seem to best predict the outcome have been identified by implementing ReliefF and Recursive Feature Elimination algorithms, upon these selected features prediction models have been developed using Logistic regression and Neural Network. The results show that Neural Network is the best predictor model for our problem with the input variables (features) selected by ReliefF algorithm. Further the features have also been ranked according to their share in predicting the outcome by use of Recursive Feature Elimination.
2 Methods
2.1 Data
Clinical data of five hundred and fifty-nine patients that have underwent treatment of chemotherapy or radiotherapy has been taken [3]. This data is available publicly at [5]. The data includes 41 features. Two-year survival is taken as the study endpoint. The data needed to go through a data cleaning step due to presence of noise (missing values). Data cleaning is a crucial step in machine learning process [6]. Mean binning method has been applied for data cleaning (i.e. to remove missing values), but new values (missing values) derived by data cleaning are guesses made based on the mean value of data, thus these values are not entirely reliable. It is necessary for data to be consistent while applying machine learning algorithms to get better results [7], so the missing values have been removed. Finally after performing the data cleaning process final dataset contains 239 patient values. Data taken for model development has the statistics shown in Table 1.
2.2 Model Development
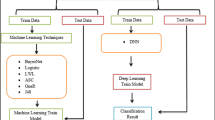
Systematic procedure is followed for model development, as shown in Fig. 1. The steps include a Data cleaning step where the data is transformed according to the suitability of the algorithm. The next step includes Feature selection, as data includes many features it is necessary to know which feature best predicts the output.
These features selected play as input to the model. Next the model is developed and trained upon these features. Finally, the model is evaluated against the training dataset. TensorFlow is used for model development.
2.2.1 Feature Selection
During the Feature Selection process a subset of features from all features are selected. It is used to make the data interpretable and insightful while reducing the dimensionality [8]. Feature selection different has methods such as filter methods, wrapper methods, embedded scheme [9]. We have implemented Recursive Feature elimination and ReliefF as our feature selectors.
2.2.1.1 ReliefF
It works by adjusting weights of features, by comparing between class and within class distance from neighbouring samples [8]. Following is a pseudo code for implementation of ReliefF [10].
-
1.
Set all weights W[A]: = 0.0;
-
2.
For i: = 1 to m do begin
-
3.
Randomly select an instance R i
-
4.
Reclassify R i instances and build a Z i dataset.
-
5.
Calculate k(Z i ) as a k percentage of the minority class in Z i.
-
6.
Find k(Z i ) nearest hit Hj and k(Z i ) near miss M j.
-
7.
For A: = 1 to a do
-
8.
\( W\left[ A \right] = W\left[ A \right] - \mathop \sum \limits_{j = 1}^{{k\left( {Z_{i} } \right)}} \frac{{Diff\left( {A,R_{i} ,H_{j} } \right)}}{{\left[ {k\left( {Z_{i} } \right)*m} \right]}} + \frac{{Diff\left( {A,R_{i} ,M_{j} } \right)}}{{\left[ {k\left( {Z_{i} } \right)*m} \right]}} \)
where W[A] denotes the weight of attributes, m is the nearest neighbours selected, Ri is a randomly selected instance from Z, Z is a set on instances, Hj and Mj are number of hits and miss \( (k\left( {Z_{i} } \right)) \). Diff function is used to calculate the difference between attribute A and instance Ri. ReliefF algorithm selected intake_who, T_stage, N_stage, Modality, CumultativeTotalTumorDose, FEV, tumorload, ott as our features.
2.2.1.2 Recursive Feature Elimination
Recursive Feature elimination is a type of wrapper method. It creates a linear SVM (support vector machine) model for each iteration, where SVM is a classification algorithm. For each iteration it works by removing the feature has the least significance, this process is repeated until all features in dataset are exhausted [11]. After all features have been removed features are ranked according to when they are removed, the later the feature is removed the higher the rank it attains. Ranking of intake_who, T_stage, N_stage, Modality, CumultativeTotalTumorDose, FEV, tumorload, ott was found to be in the order of 4, 6, 8, 2, 5, 7, 1, 3 respectively, with tumorload ranked as first.
2.2.2 Perceptron Network
The first model we have created is a linear classifier. This is simplest type of neural network, with a single-layer perceptron. It uses the equation Eq. 1 for passing values from input layer to the next layer, where wi represents the weights, xi the input attributes and b is the bias term [12].
In this network the first layer is the input units layer and second layer is both the output layer as well as hidden layer. We have taken features selected by reliefF as input units. Learning rate was set to 0.2 as it is said to be optimal. Activation function as sigmoid function and cross entropy as cost function, as it learns fast if the error is large as compared to quadratic cost function. Sigmoid function has the equation shown in Eq. 2, it was taken as cost function (i.e. z) as it maps the input in range of 0–1 which helps in classifying it to an output class. Network structure of Linear Classifier used for development of our model is shown in Fig. 2. Further it is possible to derive the derivative of logistic regression used for calculation of gradient descent, the derivative is shown in Eq. 3. Cross Entropy cost function uses equation shown in Eq. 4, which allows for faster learning, because of the fact that larger the difference the faster it learns, in Eq. 4 p is the value derived from Eq. 2 and y is the true value.
An improvement in the linear classifier is a single layer neural network with n neurons. Our implementation or Neural Network has 1 hidden layer with 10 neurons. It has a learning rate of 0.05, with rectified linear unit (ReLU) as activation function and softmax cross entropy as cost function. ReLU is based on the equation shown in Eq. 5.
It returns either a max value or 0 according to the input, i.e. 0 for negative values and the input value for positive value.
Softmax Cross Entropy is used as the cost function. Softmax is based on the equation shown in Eq. 6, it takes a N-dimensional vector and transforms it into a vector of real number in range (0, 1), where a is the value of nth vector. Derivate of softmax is shown in Eq. 7. Cross Entropy finds the distance between what the model predicted as the true value and the real true value, it is defined in Eq. 8. Structure of our Neural Network is shown in Fig. 2.
3 Results
A train test split of 90–10% is taken. Min-Max normalization is applied for data transformation. Confusion Matrix of each model has been computed for assessment of the model. It uses four variables for assessment of model. True positives (TP) depicts the positive tuples that were correctly labelled as positive by the classifier. True negative (TN) represents the negative tuples that were correctly labelled as negative by the classifier. 3. False Positives (FP) are the negative tuples that were classified as positive by classifier. 4. False Negative (FP) are the positive tuples that were falsely classified as negative.
Comparison with previously developed models for two year survival suggests Neural Network outperforming other models. SVM model has AUC of 0.59 [13], Bayesian network of Jayasurya et al. has AUC of 0.56 [14], Bayesian network of Arthur Jochems et al. has AUC of 0.66 [3]. Function corresponding for plotting a logit function is shown in Eq. 9. Output class is then decided by applying Eq. 9 to input variables and then Eq. 10 to the output of Eq. 9 get the final prediction. The t in the Eq. 10 represents the output of Eq. 9. The features selected by Feature Selection algorithm are taken as input variables in our model. This model was created for comparison purpose.
Plane created to separate the classes has a intercept of −1.7257 and coefficients for intake_who is 0.15505293, T stage is −0.23676378, N stage is 0.41042416, Modality is −0.00192756, TotalTumorDose is 0.01071944, tumorload is −0.00377834 and that of ott is 0.0210605 respectively. Accuracy of 0.6736 is achieved for this model.
Confusion Matrix for logistic regression is shown in Table 2. Table 2 shows that from total of 239 values represented as n, logistic regression identified 46 (TP) true positive out of 98 positive values (i.e. The patient survived, total actual yes), whereas 115 (TN) true negative from 141 negative values (i.e. The patient died, total actual no). While the False Negative (FN) and False Positive (FP) were found to be 52 out of 98 and 26 out of 141 respectively.
Confusion matrix for single perceptron neural network model is shown in Tabel 3. We tried to remove the features that ranked the lowest in the feature selection, expecting to get a better result, but the same accuracy was reached proving that the model assigned a negligible weight to these features. This proves that Linear Classifier weighs the features as per their importance in predicting the output.
Table 3 shows that from total of 239 values represented as n, logistic regression identified 51 (TP) true positive out of 98 positive values (i.e. The patient survived, total actual yes), whereas 110 (TN) true negative from 141 negative values (i.e. The patient died, total actual no). While the False Negative (FN) and False Positive (FP) were found to be 47 out of 98 and 31 out of 141 respectively. Comparatively Single Perceptron Neural Network is found to be better than logistic regression.
Confusion Matrix of multilayer neural network model is shown in Table 4. Different choices of artificial neurons as well as for number of hidden layers are taken. ReLU was used as the activation function with learning rate of 0.2 and one hidden layer with ten neurons. Increasing both the number of neurons and hidden layer for our model deteriorated the outcome. This proved that increasing the hidden layers or the number of neurons does not necessarily increase the outcome.
Table 2 shows that from total of 239 values represented as n, logistic regression identified 67 (TP) true positive out of 98 positive values (i.e. The patient survived, total actual yes), whereas 116 (TN) true negative from 141 negative values (i.e. The patient died, total actual no). While the False Negative (FN) and False Positive (FP) were found to be 31 out of 98 and 25 out of 141 respectively, it is found that this model performed better than any previously developed model.
Table 5 shows comparison of models developed in this study along with their measures derived from confusion matrix, where Accuracy shows how frequently is the classifier predicts correctly, Misclassification rate depicts how frequently it is false, True Positive Rate shows when the output is actually 1, how many times does the model classify it as 1, False Positive Rate shows when the output is actually 0, how frequently does the model classify it as 1, Specificity is when the output is actually 0, how frequently does it predict 0, Precision is when it predicts 1, how often is it right. Accuracy of Multilayer Neural Network is 0.7656 which is greater than other models.
4 Conclusion
This main objective of this method was to develop an accurate prediction model for two year survival prediction of patients who have suffered from non-small cell lung cancer. We propose a model to predict the two-year survival by use relief with Multilayer Neural Network. ReliefF should be used for feature selection while neural network should be adopted to develop the prediction model for two-year survival prediction of NSCLC patients. This study solves problems relating to the prediction of two-year survival of non-small cell lung cancer. Using neural network to predict two year survival of non-small cell lung cancer is a novel approach that has not been worked on which provides better result compared to previously developed models.
References
Siegel RL, Miller KD, Jemal A (2018) Cancer statistics. CA Cancer J Clin 68(1):7–30
Clément-Duchêne C, Carnin C, Guillemin F, Martinet Y (2010) How accurate are physicians in the prediction of patient survival in advanced lung cancer? Oncol Express 15(7):782–789
Jochems Arthur et al (2017) Developing and Validating a Survival Prediction Model for NSCLC Patients Through Distributed Learning Across 3 Countries. Int J Radiat Oncol *Biol* Phys 99(2):344–352
Mei X (2017) Predicting five-year overall survival in patients with non-small cell lung cancer by reliefF algorithm and random forests. In: 2017 IEEE 2nd advanced information technology, electronic and automation control conference (IAEAC)
https://www.cancerdata.org/publication/developing-and-validating-survival-prediction-model-nsclc-patients-through-distributed. Accessed Jan 2018
Devi S et al (2015) Study of data cleaning and comparision of data cleaning tools. Int J Comput Sci Mob Comput 4(3):360–370
Li X, Shi Y, Li J, Zhang P (2007) Data mining consulting improve data quality. Data Sci J 6
Wang Z et al (2016) Application of ReliefF algorithm to selecting feature sets for classification of high resolution remote sensing image. In: 2016 IEEE international geoscience and remote sensing symposium (IGARSS)
Ladha L (2011) Feature selection methods and algorithms. Int J Comput Sci Eng 3
Bolón-Canedo V, Sánchez-Maroño N, Alonso-Betanzos A (2013) A review of feature selection methods on synthetic data. Knowl Inf Syst 34
Lin G et al (2012) A support vector machine-recursive feature elimination feature selection method based on artificial contrast variables and mutual information. J Chromatogr B Anal. Technol Biomed Life Sci
Yuan LCJ, GX, HCH (2012) Recent advances of large-scale linear classification. In: Proceedings of the IEEE
Dehing-Oberije C et al (2009) Development and external validation of prognostic model for 2-year survival of non-small-cell lung cancer patients treated with chemotherapy. Int J Radiat Oncol Biol Phys 74:355–362
Jayasurya K et al (2010) Comparison of Bayesian network and support vector machine models for two-year survival prediction in lung cancer patients treated with radiotherapy. Med Phys 37:1401–1407
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Nature Switzerland AG
About this paper
Cite this paper
Dagli, Y., Choksi, S., Roy, S. (2019). Prediction of Two Year Survival Among Patients of Non-small Cell Lung Cancer. In: Peter, J., Fernandes, S., Eduardo Thomaz, C., Viriri, S. (eds) Computer Aided Intervention and Diagnostics in Clinical and Medical Images. Lecture Notes in Computational Vision and Biomechanics, vol 31. Springer, Cham. https://doi.org/10.1007/978-3-030-04061-1_17
Download citation
DOI: https://doi.org/10.1007/978-3-030-04061-1_17
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-04060-4
Online ISBN: 978-3-030-04061-1
eBook Packages: EngineeringEngineering (R0)