Abstract
Flow cytometry is a powerful analytical method, allowing to measure several properties individually for even hundreds of thousands of particles contained in some sample. Their joint distribution is a highly informative descriptor, yet directly unusable for standard machine learning methods.
Hence, such data is traditionally pre-processed into numerical features, which is often a manual or semi-automatic process. This paper introduces flowForest, an ensemble classifier capable of directly processing flow cytomtery data, modelled after the popular Random Forest method. We demonstrate that it can achieve high classification performance in a fully automatic way.
Access provided by CONRICYT-eBooks. Download conference paper PDF
Similar content being viewed by others
Keywords
1 Introduction
Flow cytometry is a modern, high-throughput laboratory technique for a thorough analysis of various particle suspensions. In a flow cytometer, the said suspension is forced though a thin capillary, in a way that only a single particle at a time passes through a system of detectors. This way, the machine can in principle analyse each particle separately, and generate a number of independent measurements corresponding to its various properties; some systems may even use this data to sort particles of certain classes into separate containers for further investigation.
Most often, the method is used to analyse suspensions of biological cells and cell fragments (usually blood), while detectors measure light scattering properties and fluorescence emissions of a number of fluorophore dies that can be selectively attached to particles based on their characteristics, usually by using modified antibodies.
Flow cytometry is widely used in biomedical research and applications; among others in agriculture [6, 15], ecology [7, 16], immunology [4, 9] and medicine [1, 13]. Still, the volume and complexity of the data it provides makes it an important field for applications of computational data analysis techniques [11, 14].
In this paper we investigate a problem of supervised classification of suspensions, each quantified by a single or a series of flow cytometry experiments. Traditionally, this is done in two stages [3, 8, 12]: first, the raw flow cytometry data is somehow, often manually, clustered in order to be reduced into a set of particle populations. Then, population cardinalities and other properties are used as features, yielding a fairly standard information system which can be used with numerous supervised classification methods.
In contrast, we aim to develop a fully automatic, generic method which would not rely on an explicit notion of particle populations, but rather some intrinsic mapping established from the training data. To this end, we propose a novel algorithm called flowForest, which is a modification of Random Forest, a popular, powerful and versatile model for classification and regression proposed by Breiman in 2001 [5].
1.1 Background
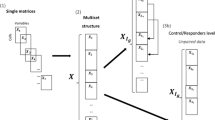
For a sake of flowForest definition, we have assumed a certain model of the information system representing a generic flow cytometry experiment; it does not cover all possibilities, however we believe it is generic enough for flowForest to be widely applicable in practice. The said information system is organised as follows; at the top level we have N objects, each corresponding to one sample and annotated with a binary decision. Either certain class can be featured or they may be equivalent, which allows for controlling on which aspects of a problem the algorithm shall focus; this aspect will be further discussed in Sect. 2.
While particles in a sample may often be stained to expose more properties than a cytometer may measure, it is common that one sample is split into T parts (further call tubes), which are then stained according to different protocols to expose various sets of properties, and finally, separately fed to a flow cytometer. To facilitate this, we assume that each object contains T flow cytometry measurements numbered from 1 to T, and that t-th measurement in each object corresponds to a sample stained according to a same protocol t, consequently that the cytometer measured a set of \(p_t\) parameters in that case.
Finally, each measurement is a set of some number of events, each of which is a vector of numbers, measured values of parameters \(p_t\). Ideally, any event should correspond to a single particle, but in practice it may as well correspond to a small group of particles, some contaminant or be a measurement artefact; hence, a modelling method must be robust to such inputs. Anyhow, \(p_t\) defines a multidimensional phase space in which events are located according to some probability density function representing a composition of various populations of particles in the sample. The flow cytometry analysis heavily relies on a concept of a gate, a compact subset of this space of an arbitrary shape which naturally describes a certain subset of events which it contains. In particular, gates are often understood strictly as corresponding to a certain particle population; yet, in this paper we will adhere to a general definition, as, quite obviously, inference on such meaningful gates is logically equivalent to an inference on a combination of seemingly nonsense gates (which is actually what flowForest does). While in general standard gate is relevant only in context of a single tube and we consider multi-tube data, it is handy to extend its notion in this regard; i.e., we will further assume that a gate also denotes on which tube it operates.
2 Materials and Methods
FlowForest, similar to Random Forest, is an ensemble classifier composed of decision trees; weak, weakly correlated and non-biased classifiers, predictions of which are agreed via simple voting. Still, flowForest uses a specialised form of decision trees—which branches operate directly on flow cytometry data rather than through a previously defined or calculated descriptors. In this regard, it can be considered to be a special case of a First Order Random Forest [17]; still, we believe that the applied specialisations are crucial for its robustness and computational feasibility. Alternatively, according to the Generalised Random Forest framework proposed in [10], flowForest is identical to Random Forest except uses pivot classifiers composed of a gate G, event statistic E, a function mapping event subset into a single real value, finally a threshold value mapping such statistic’s value into a binary outcome, L or R. Such a 3-tier structure is motivated by the flow cytometry data structure, and adheres to the variability of construction process of each tier. The threshold can be easily and explicitly optimised via simple sweep. The set of reasonable event statistics is both small and basically forced by the data structure motivating an easy best-of-few selection. On the other hand, the gate is well fitted to be selected randomly (provided certain quality requirements). Such a substantial source of randomness lowers correlations between trees, which is essential for voting to bring performance improvements.

Precisely, flowForest is built using the following procedure, presented in a pseudocode form as Algorithm 1. Every tree is grown using different N-element bootstrap sample of the training N-element set, called bag. This way roughly one-third of training objects are not used in the bootstrap sample for a given tree; they form a so-called out-of-bag subset (OOB).
The tree is built according to a standard, recursive partitioning scheme. For an input set of objects we first check if they belong to a single class; if so, a leaf with this class is generated. Otherwise, we build a split. To this end, m gates are randomly generated and for each of them and each object a table of statistics is calculated, containing the count and the average value of each of k parameters for events enclosed by the gate. For these \(m \cdot (k+1)\) constructed statistics, we consider all possible N threshold values and assess their performance of splitting the decision into possibly homogeneous parts; Gini impurity is used as a criterion. Finally, we select a single best triple of gate, statistic and threshold as the pivot, and recursively apply this procedure for each part of the split input. As in the case of the Random Forest method, we aim at growing trees to a maximum extent possible, that is the maximal depth is set large enough to be only a technical safeguard, and we do not trim the final tree.
After repeating this randomized procedure n times, a collection of n trees is obtained, constituting a flowForest. Classification of an object is done by simple voting of all trees in the ensemble; the fraction of votes on certain class is returned as well, which can be used as a continuous estimate of the model confidence.
The crucial part of this approach is the gate generation step; here we have used a simple idea of making a cuboid centred on a random event and sized randomly, though proportional to the dimensions of the cloud of all events. Naturally, we do not expect such gates to be meaningful as understood by a flow cytometry expert; they are rather probes which results become a description of event populations only after being combined by the overall model structure. Precisely, we use a following algorithm. First, we select a random tube. One event C is randomly picked from randomly chosen object from that tube. Let’s now assume that event C is characterized by k parameters \((c_1, c_2, \ldots c_k)\). Subsequently, k numbers \(e_1, e_2, \ldots , e_k\) are sampled from uniform distribution \(\mathcal {U}(0.05, 1)\). Let \(a_i\) and \(b_i\) be, respectively, 5-th and 95-th percentile of events distribution on i-th parameter. We define
for all k parameters. Finally, we construct a cuboid rectangular gate defined as follows \( \left[ c_1 - r_1, c_1 + r_1\right] \times \left[ c_2 - r_2, c_2 + r_2\right] \times \ldots \times \left[ c_k - r_k, c_k + r_k\right] . \) As mentioned earlier, not all possible random gates are fit for further consideration; some may be trivially nonsense, like being to small or unfortunately located to contain any event with reasonable odds. On the other hand, gates containing very small numbers of events (several per object on average) will have a substantial entropy and may lead to overfitting, similar to how categorical variables with a number of levels close to the number of objects would behave in a regular Random Forest model.
This is why we employ a simple quality control (QC) procedure of dropping any gate which contains less than two events for some object. One should note, however, that gates covering almost all events are not problematic, as they catch global properties of the sample. Additionally, in case there is a featured positive class, we also reject gates for which the average count of contained events is higher for the objects of the negative class than for of the positive class. This way, splits are substantially more likely to be based on the representative populations of cells for the positive class.
Obviously, it is possible that some other method of generating gates may be more appropriate, especially in certain cases; such modifications are the most natural path of extending flowForest in the future. One should also note that such a framework allows for incorporating existing expert knowledge, either by also considering gates given a priori, or by evaluating simple splits on some meta attributes. These extensions are not considered in this work, however, as we focus on formulating a possibly generic and autonomous method.
Similar to Random Forest, flowForest can establish an OOB estimate of an error. This procedure works as follows. Each object is predicted by a subset of trees for which this object was OOB, and only those predictions are combined by a regular voting. Such predictions are then compared with true classes and summarized into an OOB estimation of a prediction error, or any other possible performance metric (precision, recall, F-score, etc.).
FlowForest is implemented as an open source R package, available from https://github.com/flowforest/flowforest.
3 Results and Discussion
We have assessed the algorithm using three real-world flow cytometry data sets, earlier used in the FlowCAP II challenge [2]:
-
AML, discrimination between patients affected by acute myeloid leukaemia and healthy donors, based on blood and bone marrow aspirate samples. 359 objects (316 normal, negative, and 43 AML, positive), 8 tubes, 7 parameters per each tube, effectively 42 parameters total.
-
HVTN, discrimination between post-HIV vaccine blood samples stimulated by two HIV antigens. 96 objects (48 GAG-, 28 ENV-stimulated, equivalent), single tube, 11 parameters (we discarded the technical, non-informational time parameter, corresponding to the time of an event detection).
-
HEU, discrimination between infants exposed to HIV but uninfected and not exposed to HIV, based on blood samples. 44 objects (24 unexposed, negative, and 20 HIV-exposed, positive), 7 tubes, 10 parameters (as in case of HVTN, we discarded the time parameter) per each tube, effectively 70 parameters total.
During the challenge, all data sets were split into a training and testing subsets; we have performed our analysis both using this split, to obtain accuracy numbers comparable with those achieved by the participants. Next, we have performed a 10-fold cross-validation to investigate the robustness of the flowForest method; while classes are not balanced, we employed a slight variation of a canonical methodology, forcing all folds to have the same class distribution as the whole set. Finally, we have trained a model on a full data set to gather the OOB error estimate.
While flowForest is a stochastic algorithm, all these assessments have been replicated 10 times with different random seeds. We have investigated a number of gate tries parameter m values, namely 1, 2, 5, 10, 20, 50 and 100. The tree count parameter n was fixed to 200, which we found enough for ensemble predictions to stabilise. Performance was measured as raw accuracy, F-score and the area under ROC curve (AUROC). As the model confidence score required for AUROC, we have employed the fraction of trees in the ensemble which voted on the positive class.
The collected results for AML and HVTN problems have been presented on Fig. 1. One can see that the model was quite accurate, yielding over 90% accuracy in all investigated cases; there is also a good agreement between cross-validation and OOB estimates of error, which is important provided that the latter can be calculated in roughly one tenth of the CPU time. Both had relatively low variance across realisations, proving that the algorithm has converged and is robust to small disturbances.
Neither FlowCAP test subset seem to be representative for its corresponding full data set, however they allow us to compare flowForest with other methods, described in the after-challenge paper [2]. Using this metric, in case of the AML set, the method is able to reliably provide perfect accuracy for \(m=5\) or 10, namely achieve perfect classification in 65% of realisations, and a perfect ordering (AUROC=1) in 85%. This is consistent with the results of FlowCAP, in which 9 out of 23 submissions were perfect and the median F-score was .97; also the FlowCAP organisers concluded that the problem was relatively easy, and the only issue was a single outlier object.
On the other hand, HVTN was more challenging; flowForest required \(m=50\) to achieve best test result, and the average F-score was .95; this is worse than 6 (out of 9) methods which obtained perfect classification, and equal to one other method. However, only one of them, flowType+feaLect, was not a part of an ad hoc pipeline tuned to the problem and did not used biological knowledge.
It is also interesting to focus on \(m=1\), which corresponds to a forest built on random gates, selected only to fulfil QC criteria but not optimal with respect to the split. While, as expected, it never was an optimal value, it lead to a surprisingly good performance, showing the capabilities of a tree ensemble structure.
The HEU set was claimed to be non-informative by the challenge organisers; despite this fact, it is interesting to check whether flowForest can provide a negative answer in such case. The results of this experiment are presented on Fig. 2. One can see that in all investigated set-ups the model has degenerated and predicted all test objects as not-exposed; similarly, distributions of AUROC and accuracy under cross-validation and OOB were centred around random-guessing values.
The Fig. 3 presents the evolution of the model accuracy during the forest construction; one can see that in case of meaningful AML and HVTN problems the process converges, and that there is a significant gain from gathering trees into an ensemble, as expected from a Random Forest-based method. In case of HEU, AUROC converges to a nonsense value, and its course is substantially more variable.
4 Conclusions
The undoubtful, rising role of flow cytometry in biomedical research and medicine, as well as the volume and complexity of data it provides, calls for a novel, more automated analytical tools.
In this paper, we propose flowForest, a novel classifier based on the popular Random Forest method. We show that it can provide an accurate, robust classification on a pure flow cytometry outputs, without specialised pre-processing, injected biological knowledge, ad hoc assumptions nor any other prior information, and with minimal hyper-parameter tuning. To our knowledge, this is the only available method of a said capability, with virtually all other approaches following a two-step scenario in which data is first gated, and the inference is done only on such obtained, simplified representation. This way flowForest may discover more subtle aspects of the problem, by effectively investigating gates that are only relevant in specific, local contexts.
We also demonstrate, based on the HEU dataset, that it can reliably detect an inconclusive problem, avoiding over-fitting despite highly dimensional representation of data it uses. Quite obviously, this is a crucial property for a reliability of any potential methodology involving it, which is especially important given numerous uses of machine learning in biomedical research had suffered from such issues.
Thus, we believe it may be useful to assist manual studies, as it provides a perfectly unbiased view of data, or become a part of an automatic analysis pipeline.
References
Aebisher, D., Bartusik, D., Tabarkiewicz, J.: Laser flow cytometry as a tool for the advancement of clinical medicine. Biomed. Pharmacother. 85, 434–443 (2017)
Aghaeepour, N., et al.: Critical assessment of automated flow cytometry data analysis techniques. Nat. Methods 10(3), 228–238 (2013)
Aghaeepour, N., Nikolic, R., Hoos, H.H., Brinkman, R.R.: Rapid cell population identification in flow cytometry data. Cytom. Part A 79A(1), 6–13 (2011)
Bacher, P., Scheffold, A.: New technologies for monitoring human antigen-specific T cells and regulatory T cells by flow-cytometry. Curr. Opin. Pharmacol. 23, 17–24 (2015)
Breiman, L.: Random forests. Mach. Learn. 45, 5–32 (2001)
Czeh, A., et al.: A flow cytometry based competitive fluorescent microsphere immunoassay (CFIA) system for detecting up to six mycotoxins. J. Immunol. Methods 384(1–2), 71–80 (2012)
Dashkova, V., Malashenkov, D., Poulton, N., Vorobjev, I., Barteneva, N.S.: Imaging flow cytometry for phytoplankton analysis. Methods 112, 188–200 (2017)
Ge, Y., Sealfon, S.C.: flowpeaks: a fast unsupervised clustering for flow cytometry data via k-means and density peak finding. Bioinformatics 28(15), 2052–2058 (2012)
Kanegane, H., et al.: Flow cytometry-based diagnosis of primary immunodeficiency diseases. Allergol. Int. 67(1), 43–54 (2018)
Kursa, M.B.: Generalised random forest space overview (2015). https://arxiv.org/abs/1501.04244
Lizard, G.: Flow cytometry analyses and bioinformatics: interest in new softwares to optimize novel technologies and to favor the emergence of innovative concepts in cell research. Cytom. A 71A, 646–647 (2007)
Lo, K., Hahne, F., Brinkman, R.R., Gottardo, R.: flowclust: a bioconductor package for automated gating of flow cytometry data. BMC Bioinform. 10(1), 1–8 (2009)
Maguire, O., Tario, J.D., Shanahan, T.C., Wallace, P.K., Minderman, H.: Flow cytometry and solid organ transplantation: a perfect match. Immunol. Investig. 43, 756–774 (2014)
O’Neill, K., Aghaeepour, N., Špidlen, J., Brinkman, R.: Flow cytometry bioinformatics. PLoS Comput. Biol. 9, 1–10 (2013)
Sliwinska, E.: Flow cytometry - a modern method for exploring genome size and nuclear DNA synthesis in horticultural and medicinal plant species. Folia Hortic. 30(1), 103–128 (2018)
Takahashi, T.: Life cycle analysis of endosymbiotic algae in an endosymbiotic situation with paramecium bursaria using capillary flow cytometry. Energies 10(9), 1413 (2017)
Van Assche, A., Vens, C., Blockeel, H., Džeroski, S.: First order random forests: Learning relational classifiers with complex aggregates. Mach. Learn. 64(1), 149–182 (2006)
Acknowledgements
This work has been financed by the National Science Centre, grant 2011/01/N/ST6/07035. Computations were performed in ICM, grant G48-6.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Nature Switzerland AG
About this paper
Cite this paper
Piotrowski, B.P., Kursa, M.B. (2018). Fully Automatic Classification of Flow Cytometry Data. In: Ceci, M., Japkowicz, N., Liu, J., Papadopoulos, G., Raś, Z. (eds) Foundations of Intelligent Systems. ISMIS 2018. Lecture Notes in Computer Science(), vol 11177. Springer, Cham. https://doi.org/10.1007/978-3-030-01851-1_1
Download citation
DOI: https://doi.org/10.1007/978-3-030-01851-1_1
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-030-01850-4
Online ISBN: 978-3-030-01851-1
eBook Packages: Computer ScienceComputer Science (R0)