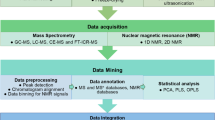
Abstract
Metabolomics is one of the most recent additions to the functional genomics approaches. It involves the use of analytical chemistry techniques to provide high-density data of metabolic profiles. Data is then analyzed using advanced statistics and databases to extract biological information, thus providing the metabolic phenotype of an organism. Large variety of metabolites produced by plants through the complex metabolic networks and their dynamic changes in response to various perturbations can be studied using metabolomics. Here, we describe the basic features of plant metabolic diversity and analytical methods to describe this diversity, which includes experimental workflows starting from experimental design, sample preparation, hardware and software choices, combined with knowledge extraction methods. Finally, we describe a scenario for using these workflows to identify differential metabolites and their pathways from complex biological samples.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Hartmann T (2007) From waste products to ecochemicals: fifty years research of plant secondary metabolism. Phytochemistry 68:2831–2846
Vranova E, Coman D, Gruissem W (2012) Structure and dynamics of the isoprenoid pathway network. Mol Plant 5:318–333
Zhang P, Foerster H, Tissier CP, Mueller L, Paley S, Karp PD, Rhee SY (2005) MetaCyc and AraCyc. Metabolic pathway databases for plant research. Plant Physiol 138:27–37
Plant Metabolic Network. http://www.plantcyc.org/release_notes/content_statistics.faces. Accessed 02-Feb-2013
Wink M (2010) Introduction: biochemistry, physiology and ecological functions of secondary metabolites. Annu Plant Rev 40: Biochemistry of Plant Secondary Metabolism
Rhodes M (1994) Physiological roles for secondary metabolites in plants: some progress, many outstanding problems. Plant Mol Biol 24:1–20
Sakakibara H (2006) Cytokinins: activity, biosynthesis, and translocation. Annu Rev Plant Biol 57:431–449
Fluck RA, Leber PA, Lieser JD, Szczerbicki SK, Varnes JG, Vitale MA, Wolfe EE (2000) Choline conjugates of auxins. I. Direct evidence for the hydrolysis of choline-auxin conjugates by pea cholinesterase. Plant Physiol Biochem 38:301–308
Winkel-Shirley B (2001) Flavonoid biosynthesis. A colorful model for genetics, biochemistry, cell biology, and biotechnology. Plant Physiol 126:485–493
Winkel-Shirley B (2002) Biosynthesis of flavonoids and effects of stress. Curr Opin Plant Biol 5:218–223
Winkel-Shirley B (2001) It takes a garden. How work on diverse plant species has contributed to an understanding of flavonoid metabolism. Plant Physiol 127:1399–1404
Narasimhan K, Basheer C, Bajic VB, Swarup S (2003) Enhancement of plant-microbe interactions using a rhizosphere metabolomics-driven approach and its application in the removal of polychlorinated biphenyls. Plant Physiol 132:146–153
Rasmussen S, Parsons AJ, Jones CS (2012) Metabolomics of forage plants: a review. Ann Bot 110:1281–1290
Reuben S, Bhinu VS, Swarup S (2008) Rhizosphere metabolomics: methods and applications. In: Karlovsky P (ed) Secondary metabolites in soil ecology. Springer, Berlin, pp 37–68
Schenk PM, Kazan K, Wilson I, Anderson JP, Richmond T, Somerville SC, Manners JM (2000) Coordinated plant defense responses in Arabidopsis revealed by microarray analysis. Proc Natl Acad Sci U S A 97:11655–11660
Dalmolin RJ, Castro MA, Rybarczyk Filho JL, Souza LH, de Almeida RM, Moreira JC (2011) Evolutionary plasticity determination by orthologous groups distribution. Biol Direct 6:22
Hart Y, Mayo AE, Milo R, Alon U (2011) Robust control of PEP formation rate in the carbon fixation pathway of C(4) plants by a bi-functional enzyme. BMC Syst Biol 5:171
Angelovici R, Fait A, Zhu X, Szymanski J, Feldmesser E, Fernie AR, Galili G (2009) Deciphering transcriptional and metabolic networks associated with lysine metabolism during Arabidopsis seed development. Plant Physiol 151:2058–2072
Brown PD, Tokuhisa JG, Reichelt M, Gershenzon J (2003) Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62:471–481
Desbrosses GG, Kopka J, Udvardi MK (2005) Lotus japonicus metabolic profiling. Development of gas chromatography-mass spectrometry resources for the study of plant-microbe interactions. Plant Physiol 137: 1302–1318
Bhalla R, Narasimhan K, Swarup S (2005) Metabolomics and its role in understanding cellular responses in plants. Plant Cell Rep 24:562–571
Alisdair RF, Richard NT, Arno JK, Lothar W (2004) Innovation: metabolite profiling: from diagnostics to systems biology. Nat Rev Mol Cell Biol 5
De Vos R, Moco S, Lommen A, Keurentjes J, Bino R, Hall R (2007) Untargeted large-scale plant metabolomics using liquid chromatography coupled to mass spectrometry. Nat Protoc 2:778–791
Hall R (2006) Plant metabolomics: from holistic hope, to hype, to hot topic. New Phytol 169:453–468
Glinski M, Weckwerth W (2006) The role of mass spectrometry in plant systems biology. Mass Spectrom Rev 25:173–214
Patti G (2011) Separation strategies for untargeted metabolomics. J Sep Sci 34:3460–3469
Werner E, Croixmarie V, Umbdenstock T, Ezan E, Chaminade P, Tabet JC, Junot C (2008) Mass spectrometry-based metabolomics: accelerating the characterization of discriminating signals by combining statistical correlations and ultrahigh resolution. Anal Chem 80:4918–4932
Kueger S, Steinhauser D, Willmitzer L, Giavalisco P (2012) High-resolution plant metabolomics: from mass spectral features to metabolites and from whole-cell analysis to subcellular metabolite distributions. Plant J 70:39–50
Kim HK, Choi YH, Verpoorte R (2011) NMR-based plant metabolomics: where do we stand, where do we go? Trends Biotechnol 29:267–275
Dunn WB, Bailey NJ, Johnson HE (2005) Measuring the metabolome: current analytical technologies. Analyst 130:606–625
Morita A, Horie H, Fujii Y, Takatsu S, Watanabe N, Yagi A, Yokota H (2004) Chemical forms of aluminum in xylem sap of tea plants (Camellia sinensis L.). Phytochemistry 65:2775–2780
Veenstra T (2012) Metabolomics: the final frontier? Genome Med 4:40
David SW (2008) Quantitative metabolomics using NMR. TrAC Trends Anal Chem 27
Bedair M, Sumner LW (2008) Current and emerging mass-spectrometry technologies for metabolomics. Trac-Trend Anal Chem 27:238–250
Dettmer K, Aronov P, Hammock B (2007) Mass spectrometry-based metabolomics. Mass Spectrom Rev 26:51–78
Parab GS, Rao R, Lakshminarayanan S, Bing YV, Moochhala SM, Swarup S (2009) Data-driven optimization of metabolomics methods using rat liver samples. Anal Chem 81:1315–1323
Broeckling CD, Huhman DV, Farag MA, Smith JT, May GD, Mendes P, Dixon RA, Sumner LW (2005) Metabolic profiling of Medicago truncatula cell cultures reveals the effects of biotic and abiotic elicitors on metabolism. J Exp Bot 56:323–336
Hall R, Beale M, Fiehn O, Hardy N, Sumner L, Bino R (2002) Plant metabolomics: the missing link in functional genomics strategies. Plant Cell 14:1437–1440
Bleeker PM, Diergaarde PJ, Ament K, Guerra J, Weidner M, Schutz S, de Both MT, Haring MA, Schuurink RC (2009) The role of specific tomato volatiles in tomato-whitefly interaction. Plant Physiol 151:925–935
Mayer F, Takeoka GR, Buttery RG, Whitehand LC, Naim M, Rabinowitch HD (2008) Studies on the aroma of five fresh tomato cultivars and the precursors of cis- and trans-4,5-epoxy-(E)-2-decenals and methional. J Agric Food Chem 56:3749–3757
Shuman JL, Cortes DF, Armenta JM, Pokrzywa RM, Mendes P, Shulaev V (2011) Plant metabolomics by GC-MS and differential analysis. Methods Mol Biol 678:229–246
Allwood J, Goodacre R (2010) An introduction to liquid chromatography-mass spectrometry instrumentation applied in plant metabolomic analyses. Phytochem Anal 21:33–47
Nordstrom A, Want E, Northen T, Lehtio J, Siuzdak G (2008) Multiple ionization mass spectrometry strategy used to reveal the complexity of metabolomics. Anal Chem 80:421–429
Bin Z, Jun Feng X, Leepika T, Habtom WR (2012) LC-MS-based metabolomics. Mol Biosyst 8
Obata T, Fernie A (2012) The use of metabolomics to dissect plant responses to abiotic stresses. Cell Mol Life Sci 69:3225–3243
Lei Z, Huhman D, Sumner L (2011) Mass spectrometry strategies in metabolomics. J Biol Chem 286:25435–25442
Gibon Y, Rolin D (2012) Aspects of experimental design for plant metabolomics experiments and guidelines for growth of plant material. Methods Mol Biol (Clifton, NJ) 860: 13–30
Leek JT, Scharpf RB, Bravo HC, Simcha D, Langmead B, Johnson WE, Geman D, Baggerly K, Irizarry RA (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11:733–739
Fiehn O, Wohlgemuth G, Scholz M, Kind T, Lee do Y, Lu Y, Moon S, Nikolau B (2008) Quality control for plant metabolomics: reporting MSI-compliant studies. Plant J 53:691–704
Members MSIB, Sansone SA, Fan T, Goodacre R, Griffin JL, Hardy NW, Kaddurah-Daouk R, Kristal BS, Lindon J, Mendes P, Morrison N, Nikolau B, Robertson D, Sumner LW, Taylor C, van der Werf M, van Ommen B, Fiehn O (2007) The metabolomics standards initiative. Nat Biotechnol 25:846–848
Boccard J, Veuthey JL, Rudaz S (2010) Knowledge discovery in metabolomics: an overview of MS data handling. J Sep Sci 33:290–304
Hendriks MMWB, van Eeuwijk FA, Jellema RH, Westerhuis JA, Reijmers TH, Hoefsloot HCJ, Smilde AK (2011) Data-processing strategies for metabolomics studies. Trac-Trend Anal Chem 30:1685–1698
Eliasson M, Rannar S, Trygg J (2011) From data processing to multivariate validation–essential steps in extracting interpretable information from metabolomics data. Curr Pharm Biotechnol 12:996–1004
Biswas A, Mynampati KC, Umashankar S, Reuben S, Parab G, Rao R, Kannan VS, Swarup S (2010) MetDAT: a modular and workflow-based free online pipeline for mass spectrometry data processing, analysis and interpretation. Bioinformatics 26:2639–2640
Smith CA, Want EJ, O'Maille G, Abagyan R, Siuzdak G (2006) XCMS: processing mass spectrometry data for metabolite profiling using nonlinear peak alignment, matching, and identification. Anal Chem 78:779–787
Pluskal T, Castillo S, Villar-Briones A, Oresic M (2010) MZmine 2: modular framework for processing, visualizing, and analyzing mass spectrometry-based molecular profile data. BMC Bioinformatics 11:395
Xia J, Mandal R, Sinelnikov IV, Broadhurst D, Wishart DS (2012) MetaboAnalyst 2.0—a comprehensive server for metabolomic data analysis. Nucleic Acids Res 40:W127–W133
Hegeman A (2010) Plant metabolomics—meeting the analytical challenges of comprehensive metabolite analysis. Brief Funct Genomics 9:139–148
Fan TW-M (2012) Considerations of sample preparation for metabolomics investigation. In: Fan TW-M, Lane AN, Higashi RM (eds) The handbook of metabolomics, vol 17, Methods in pharmacology and toxicology. Humana, Totowa, NJ, pp 7–27
American Society for Mass Spectrometry (2009) Metabolomics ASMS Workshop Survey 2009. http://fiehnlab.ucdavis.edu/staff/kind/Metabolomics-Survey-2009
Kind T, Fiehn O (2007) Seven Golden Rules for heuristic filtering of molecular formulas obtained by accurate mass spectrometry. BMC Bioinformatics 8:105
Biswas A, Rao R, Umashankar S, Mynampati KC, Reuben S, Parab G, Swarup S (2011) datPAV—an online processing, analysis and visualization tool for exploratory investigation of experimental data. Bioinformatics 27:1585–1586
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28:27–30
Kanehisa M, Goto S, Sato Y, Furumichi M, Tanabe M (2012) KEGG for integration and interpretation of large-scale molecular data sets. Nucleic Acids Res 40:D109–D114
Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M (2008) KEGG Atlas mapping for global analysis of metabolic pathways. Nucleic Acids Res 36:W423–W426
Sana TR, Roark JC, Li X, Waddell K, Fischer SM (2008) Molecular formula and METLIN Personal Metabolite Database matching applied to the identification of compounds generated by LC/TOF-MS. J Biomol Technol 19:258–266
ChemSpider. http://www.chemspider.com/
Horai H, Arita M, Kanaya S, Nihei Y, Ikeda T, Suwa K, Ojima Y, Tanaka K, Tanaka S, Aoshima K, Oda Y, Kakazu Y, Kusano M, Tohge T, Matsuda F, Sawada Y, Hirai MY, Nakanishi H, Ikeda K, Akimoto N, Maoka T, Takahashi H, Ara T, Sakurai N, Suzuki H, Shibata D, Neumann S, Iida T, Tanaka K, Funatsu K, Matsuura F, Soga T, Taguchi R, Saito K, Nishioka T (2010) MassBank: a public repository for sharing mass spectral data for life sciences. J Mass Spectrom 45:703–714
Kessner D, Chambers M, Burke R, Agus D, Mallick P (2008) ProteoWizard: open source software for rapid proteomics tools development. Bioinformatics 24:2534–2536
R Core Team (2012) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria
Sugimoto M, Kawakami M, Robert M, Soga T, Tomita M (2012) Bioinformatics tools for mass spectroscopy-based metabolomic data processing and analysis. Curr Bioinformatics 7:96–108
Mochida K, Shinozaki K (2010) Genomics and bioinformatics resources for crop improvement. Plant Cell Physiol 51:497–523
Chagoyen M, Pazos F (2012) Tools for the functional interpretation of metabolomic experiments. Brief Bioinformatics. doi: 10.1093 /bib/bbs055
Tokimatsu T, Sakurai N, Suzuki H, Ohta H, Nishitani K, Koyama T, Umezawa T, Misawa N, Saito K, Shibata D (2005) KaPPA-view: a web-based analysis tool for integration of transcript and metabolite data on plant metabolic pathway maps. Plant Physiol 138:1289–1300
Junker BH, Klukas C, Schreiber F (2006) VANTED: a system for advanced data analysis and visualization in the context of biological networks. BMC Bioinformatics 7:109
Letunic I, Yamada T, Kanehisa M, Bork P (2008) iPath: interactive exploration of biochemical pathways and networks. Trends Biochem Sci 33:101–103
Xia J, Wishart DS (2010) MetPA: a web-based metabolomics tool for pathway analysis and visualization. Bioinformatics 26:2342–2344
Acknowledgements
We acknowledge the financial support from the Singapore-Peking-Oxford Research Enterprise, COY-15-EWI-RCFSA/N197-1. We gratefully acknowledge Agilent Technologies, Singapore for their support in acquiring and analyzing the mass spectrometry data for the differential analysis of metabolites.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Rai, A., Umashankar, S., Swarup, S. (2013). Plant Metabolomics: From Experimental Design to Knowledge Extraction. In: Rose, R. (eds) Legume Genomics. Methods in Molecular Biology, vol 1069. Humana Press, Totowa, NJ. https://doi.org/10.1007/978-1-62703-613-9_19
Download citation
DOI: https://doi.org/10.1007/978-1-62703-613-9_19
Published:
Publisher Name: Humana Press, Totowa, NJ
Print ISBN: 978-1-62703-612-2
Online ISBN: 978-1-62703-613-9
eBook Packages: Springer Protocols