Abstract
MicroRNAs (miRNAs) are a class of small, noncoding, and single-stranded RNAs that play a key role in regulating gene expressions in most eukaryotes. Human genome encodes over 1,600 different miRNA loci, and miRNAs directly regulate expression of over 60% of protein-coding genes. The canonical miRNA biogenesis pathway has been well documented with key players identified by the efforts of many biochemical research groups. However, we still lack a mechanistic understanding of the target recognition and gene silencing processes. Over the recent years, structural, biochemical, and single-molecule biophysical studies have revealed the complexity in the interactions between miRNA and the effector protein Argonaute (Ago) as well as between Ago and the target mRNA. In this chapter, we present these findings that significantly enhanced our knowledge of the assembly and regulation by Ago–miRNA complexes.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
One of the key posttranscriptional gene regulatory mechanisms in eukaryotes is mediated by small regulatory RNAs such as microRNAs (miRNAs). miRNAs are ~22 nucleotides (nt) long, small noncoding RNAs that induce translational repression and degradation of mRNAs that are complementary to seed sequences of the miRNA (reviewed in [1, 2]). A summary of miRNA biogenesis process is presented in Fig. 8.1. Briefly, miRNA biogenesis begins with the transcription of the miRNA gene by RNA polymerase II [3,4,5,6]. A cluster of miRNAs is transcribed together as a long polycistronic transcript known as primary miRNA (pri-miRNA), which folds back on itself to form multiple hairpin structures in a single transcript (Fig. 8.1). These hairpins undergo endonucleolytic cleavage by RNase III-type enzyme Drosha in a complex with DiGeorge syndrome critical region gene 8 (DGCR8) [7,8,9,10,11]. The complex, known as the microprocessor, recognizes the junction between the hairpin structure and the single-stranded RNA and cleaves the RNA ~11 bases away from the junction [9, 12, 13]. More recently, structural and biochemical investigations have identified the molar composition of the microprocessor (one molecule of Drosha and two molecules of DGCR8), Drosha-binding motif in the basal segment of the pri-miRNA, as well as DGCR8-binding motif in the hairpin region of the RNA [13,14,15].
Microprocessor cleaves pri-miRNAs and releases ~65–70 nt long stem-loop structured RNAs known as precursor miRNAs (pre-miRNAs). Pre-miRNAs are then exported to the cytoplasm by Exportins including Exportin-5 where they are recognized by another RNase III-type enzyme Dicer [16]. Dicer recognizes both the phosphate group at the 5′ end and the 2 nt overhang structure of the pre-miRNA and cleaves the RNA ~22 nt from the ends [17,18,19]. The resulting miRNA duplex is then loaded onto Argonaute (Ago) family of proteins which discards one of the strands (known as the passenger strand) and retains the other strand (known as the guide strand). Ago–miRNA complex constitutes the core of the RNA-induced silencing complex (RISC) and uses the miRNA seed sequences as the guide to search for target mRNAs to induce posttranscriptional gene silencing [20, 21].
Numerous studies analyzed the Ago–miRNA and RISC–mRNA interactions using biochemical and biophysical single-molecule approaches. Their experimental findings were further complemented by the structural knowledge of Ago and RISC. Together, these studies have significantly advanced our understanding of the mechanism of gene regulation mediated by miRNAs. In this chapter, we present these studies.
2 Functional Domains of Ago
The overall structure of Ago family of proteins is a bilobate architecture that consists of four distinct domains: the N-terminal, PAZ, MID, and Piwi domains (Fig. 8.2) [22, 23]. Biological functions of these domains are summarized in Table 8.1. The N-terminal region forms one lobe with the PAZ domain. The function of the N-terminal region is unclear, but it may assist in the release of the target mRNA by disrupting its base pairing with the miRNA [24]. The PAZ domain can be subdivided into two subdomains separated by threonine 667; one domain consists mostly of aromatic residues, while the other subdomain folds into a structure similar to oligonucleotide/oligosaccharide binding (or OB-fold) structure that is capable of binding to single-stranded nucleic acids [22, 25,26,27]. The possibility of the interaction between the PAZ domain and the single-stranded nucleic acids is confirmed via crystallographic studies and biochemical experiments where the PAZ domain binds to single-stranded RNAs, although with low affinity [28,29,30].

The figure is adapted from [22] with American Association for the Advancement of Science, Copyright 2012
A schematic of different functional domains (top) and the ternary structure of human Ago2 (bottom).
The PAZ domain can interact and anchor the 3′ end of the miRNA [31]. The anchoring incurs steric hindrance and prevents the interaction between the last few nucleotides of the miRNA with its target mRNA. This reduces the degree of interaction between the miRNA and the target mRNA, facilitating the target release and allowing RISC to act as a multi-turnover complex [31, 32]. Furthermore, anchoring of the 3′ end of the miRNA is important for the loading of miRNA duplex onto Ago. Dicer cleavage product (miRNA duplex) contains two nucleotide 3′ overhangs which is a common characteristic of RNase type-III enzyme products [33]. This recognition of the 3′ overhang allows Ago to distinguish miRNA duplex from other small RNAs such as degradation by-products or small duplex RNAs that are derived from non-related pathways [34,35,36].
The human genome encodes four paralogs of Ago proteins (hAgo1–4). While all four proteins share the characteristic domains of the Ago family, only hAgo2 shows target cleavage activity, which is mediated by the Piwi domain [21, 37]. This domain has an RNaseH-like fold and is responsible for the endonucleolytic activity of the protein. RNaseH is an endonuclease that recognizes DNA–RNA hybrid and cleaves RNA using DNA as the template. The catalytic activity of RNaseH requires a conserved Asp-Asp-Glu/Asp motif in the catalytic center and two divalent metal ions [38]. The Piwi domain of cleavage competent Agos including hAgo2 has a very similar motif (Asp-Asp-Asp/Glu/His/Lys) [23]. Mutagenesis of this region resulted in the loss of the catalytic activity [23]. In addition, these Agos require divalent metal ions to induce RNA cleavage [21, 39, 40]. Moreover, the products of Agos and RNaseH both show 3′-OH and 5′-phosphate groups, suggesting that the two proteins induce RNA cleavage in a similar manner [21, 40, 41].
Unlike hAgo2, other three paralogs of human Agos (hAgo1, hAgo3, and hAgo4) do not show slicing activity. Examination of their Piwi domains reveals that the RNaseH-like motif in hAgo1 and hAgo4 does not match the consensus sequence and hence accounts for their inability to cleave target mRNAs. Human Ago3 shows Asp-Asp-His consensus sequence which matches the one from hAgo2, yet studies reported that hAgo3 does not show RNA cleavage activity [42]. Therefore, simple RNaseH fold structure may not account for the action mechanism of Agos.
One possible explanation is the difference in the target cleavage efficiency. In Drosophila, two Agos (Ago1 and Ago2) have the identical consensus motif, but Ago1 shows much higher cleavage efficiency than that of Ago2 due to faster target release kinetics [43]. Applying similar logic to the human Agos, hAgo3 may have much slower dissociation kinetics with the target compared to that of hAgo2, which can make hAgo3 effectively a single turnover enzyme and show much lower cleavage efficiency. Through a series of biochemical experiments using recombinant hAgo2 and hAgo3, Park et al. showed that hAgo3 loaded with miR-20a can cleave target mRNAs [44]. However, when incubated with other miRNAs such as let-7a, miR-19b, or miR-16, recombinant hAgo3 failed to induce target cleavage [44]. The authors attributed this phenomenon to the differences in the miRNA-target interaction channel between hAgo2 and hAgo3, indicating that hAgo3 has more strict substrate requirement in addition to simple sequence complementarity in order to induce target cleavage [44]. As Ago protein structure plays a key role in the miRNA-target interaction as well as during target dissociation from RISC (see below for details), the difference in action mechanism of hAgo2 and hAgo3 may arise from the differences in their interaction with the target rather than in the RNaseH-like motif in their Piwi domains. Together, these evidences call for more detailed investigation of miRNA-target interaction in combination with information on Ago protein structures to better understand the action mechanism of miRNAs.
For both cleavage competent and incompetent Agos, the Piwi domain plays an essential role in substrate recognition. While the 3′ end of the miRNA is recognized by the PAZ domain, the 5′-phosphate of the miRNA is anchored at the interface between the Piwi and the MID domains [42, 45]. Biochemical studies further showed that a divalent cation binds to this interface and interacts with the 5′-phosphate of the miRNA [28]. Furthermore, the preferential nucleotide is shown to be uridine although the effect of the nucleotide identity and the efficiency of Ago loading in cell need further investigation [46].
The main silencing effect by RISC is mediated not by target cleavage, but mostly through translational repression and subsequent RNA degradation via deadenylation and decapping [42]. The MID domain holds the key to explain the latter function of Agos as hAgo2 contains an MC motif which shows high homology to the cap-binding motif of the eukaryotic initiation factor 4E (eIF4E) [47]. Biochemical studies further showed that the MID domain can bind to the cap of the mRNA, and this interaction is required for efficient translational repression [48]. As hAgo1, hAgo3, and hAgo4 are not capable of inducing target cleavage, the cap-binding ability provided by the MID domain may be critical for these Agos to induce repressive effects. However, the MID domains of these cleavage incompetent Agos do not show the motif homologous to that of eIF4E and the exact mechanism of the cap-binding ability of these Agos remains to be investigated. Perhaps, this process may be mediated by Ago-interacting proteins that are components of RISC [49,50,51]. In addition, it remains unclear how the MID domain and Ago induce deadenylation and decapping of the target mRNA. Recently, it has been shown that targeting by miRNAs induces uridylation of the mRNA at the end of its poly(A) tail which facilitates RNA turnover [52]. Considering that mRNA turnover is responsible for most of the gene silencing effect by RISC, the role of MID domain and its cap-binding ability to induce deadenylation and decapping needs further investigation in the future.
3 Assembly of Ago–MiRNA Complex
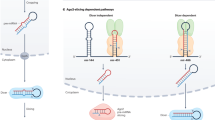
During the posttranscriptional regulation by miRNAs, miRNAs use their seed sequences to guide the RISC to find the target mRNAs while Ago functions as the effector protein of the complex [53,54,55]. The interaction between miRNAs and Agos begins with the assembly of the RISC. Since the mechanism of RISC assembly has been key aspects in understanding miRNA-mediated gene silencing, it was under extensive investigation over the past few decades. RISC assembly begins with the loading of duplex Dicer cleavage product onto Ago protein. This process is most thoroughly studied using Drosophila Ago2, but in this chapter, we will focus on the mammalian system (Fig. 8.3).

The figure is modified from [66] with Elsevier, Copyright 2012
RISC assembly in mammals. Pre-miRNAs are first loaded onto miRNA RISC loading complex (miRLC). However, the transfer mechanism of Dicer cleavage products to Ago and the function of the direct interaction between pre-miRNAs and Ago remain to be investigated.
One of the key questions in RISC assembly is how Dicer releases its cleavage product and delivers it to Ago. Numerous studies have suggested that Dicer together with Ago and TAR RNA-binding protein (TRBP) forms miRNA RISC loading complex (miRLC) which plays a key role in loading of the miRNA duplex (Fig. 8.3). In this conventional model, Dicer product is oriented by Dicer–TRBP heterodimer and is handed over to Ago in the miRLC [56,57,58]. This is further supported by the EM data where Ago is bound to miRNA duplex in complex with Dicer and TRBP [59].
However, accumulating in vitro biochemical evidences suggests that Dicer and miRLC may not be required for miRNA loading [60,61,62,63]. Using Dicer knockout embryonic stem cells, it has been shown that the loading of small RNA duplexes to Ago can occur in the absence of Dicer [64]. In addition, Ago can directly bind to pre-miRNAs and form a complex called miRNA deposit complex (miPDC; Fig. 8.3). For specific miRNAs such as miR-451 whose pre-miRNA is too short for Dicer processing, miPDC formation is responsible for mature miRNA biogenesis [65]. In other cases, miPDC can incorporate Dicer and TRBP to form miRLC and deliver pre-miRNA to Dicer for RNA processing [66].
Interestingly, it has been shown that dissociation of Ago from miRLC to form RISC with mature miRNA requires catalytically active Dicer [57]. This supports the conventional model where Ago is present as a complex with Dicer, and Dicer physically hands over its cleavage product, the miRNA duplex, to Ago [57]. Furthermore, the loading of miRNA duplex may trigger conformational change on Ago such that it can now dissociate from miRLC to form mature RISC [60, 67]. However, since small duplex RNAs can be loaded on to Ago without Dicer, it still remains unclear how the miRNA RISC loading occurs. In the end, there exist two models: Dicer product is released to the bulk solution and then loaded onto Ago in the vicinity, and Dicer is physically handing over the cleaved product to the Ago protein [66]. In either of the two cases, the interaction between Ago and Dicer and the formation of miRLC are required in order to minimize the searching process of Ago to find duplex miRNAs [58, 68]. Further investigation is required to elucidate the in-depth mechanism of Ago loading during miRNA biogenesis in mammals.
4 Target Recognition by Minimal RISC
Once the RISC assembly has been completed, Ago uses the guide strand of the miRNA embedded within the complex to search for its target mRNAs to induce posttranscriptional regulation [69, 70]. During the process, Ago searches for mRNAs whose 3′ UTR sequences are complementary to the miRNAs’ seed sequences, 2–7 or 2–8 nt from the 5′ end of the miRNA [42]. The importance of the miRNA seed sequences has been demonstrated by numerous high-throughput sequencing studies. They showed that when expression of a given miRNA is perturbed, levels of mRNAs that contain sequences complementary to seed sequence of the miRNA are significantly affected [71,72,73,74]. Detailed examination of the interaction between miRNA and its target mRNA revealed that the two RNAs hybridize in a stepwise process that is accompanied by structural changes in Ago [75].
miRNA loading and target mRNA recognition by RISC can be subdivided into five steps. The first step is the loading of the miRNA onto Ago and its effect on the accessibility of the individual nucleotides. The 5′-monophosphate and the first base of the miRNA are anchored at the interface between the MID and the Piwi domains, and the 3′ end of the miRNA is recognized by the PAZ domain of Ago [25, 28, 29, 31, 45, 76]. This anchoring of miRNA guide strand makes the first nucleotide inaccessible and opens up the seed region to the media [28, 45]. Therefore, the seed region becomes the first nucleotides that can interact with mRNAs and play an essential role in target selection by the miRNA.
Although the seed region is crucial for target recognition, not all seven bases hybridize with their complementary bases on the target mRNA simultaneously. The latter part of the seed region is inaccessible to the media due to steric hindrance imposed by the Ago protein (Fig. 8.4) [75]. Initially, only the second–fifth positions of the guide RNA are exposed and are able to interact with the target mRNAs [75]. These sequences are known as the sub-seed sequences and are responsible for the weak recognition of the target by RISC. When RISC finds an mRNA with complementary match to the sub-seed sequences, the hybridization between miRNA and mRNA is stabilized by Ago protein which assists the two to form into an A-form helix [75, 77]. This pre-organization of miRNAs to base pair only the sub-seed region greatly accelerates the target finding speed by increasing the on-rate as much as 250-folds [78].

The figure is adapted from [77] with Elsevier, Copyright 2017
In RISC, the access of the latter half of the seed region (5th–7th nt) of the miRNA is blocked by the helix-7 motif of Ago.
The initial interaction between the sub-seed sequences and the mRNAs has been supported by a number of single-molecule studies. For example, Salomon et al. designed an experiment where they introduced a series of di-nucleotide changes on mRNAs and measured the dissociation rate of the RISC–mRNA complex [78]. They found that mismatches in the first two sequences of the seed region had the greatest effects on the target binding rates compared to mismatches in other regions [78]. Interestingly, mismatches in the last two sequences of the seed had significantly weaker effects on target binding rate [78]. Similarly, Chandradoss et al. compared the binding rate of mRNA with full seed complementarity and one with partial seed complementarity. They found that the first three nucleotides of the seed region are critical for the target recognition and the latter part of the seed region did not have significant effects on the binding rate between the RISC and the target mRNA [79]. Of note, while the structural study subclassified 2–5 nt as sub-seed sequences, these follow-up single-molecule studies showed that only the 2–4 nt may act as the “mini-seed” region [75, 78, 79]. These studies support the notion that RISC uses the first three or maybe four nucleotides of the seed region for the initial target search.
The hypothesis of the existence and the significance of the sub-seed match are further supported by structural analysis of Ago protein in complex with guide miRNA. Without interaction with the target, only the second–fifth positions rather than the entire seed region are exposed to the media [22, 45, 80, 81]. This is because Ago protein induces structural constraint and makes the guide kink away from the A-form helix, in particular at the position 7. With this conformation, mRNA will not be able to hybridize and form duplex RNA beyond the fifth position of the miRNA (Fig. 8.4) [77].
This structural constraint at the position 7 is relieved by the initial interaction between the mRNA and the sub-seed sequences of the guide miRNA. The hybridization triggers structural change on Ago such that it undergoes 4 Å displacement at the region around the position 7, allowing the seventh nucleotide to adapt A-helix configuration (Fig. 8.5) [77]. This allows the sixth–eighth positions of the guide RNA to base pair with the target mRNA [77].

The figure is adapted from [75] with American Association for the Advancement of Science, Copyright 2014
Base pairing in the sub-seed region induces conformational change such that the helix-7 is shifted by 4 Å which allows further base pairing with the target.
The conformational change of Ago and the subsequent extension of hybridization between the miRNA and the target mRNA are supported by a number of single-molecule studies. They showed that there is a sharp increase in the binding affinity when the number of seed matches is increased from six to seven [78, 79]. This result is consistent with the idea that the sub-seed interaction induces changes in Ago structure such that the sixth–eighth position of the guide RNA has become accessible to hybridize with the target mRNA [82]. Without such change, these positions of the seed region will not be able to hybridize with the mRNA and thus complementarity in these bases will not affect the binding affinity with the target.
The combination of structural, biochemical, and biophysical single-molecule experiments provides a powerful approach in understanding RISC–mRNA interaction. Together, these studies converge on the idea that the target recognition by RISC is a multi-step process (Fig. 8.6). First, RISC anchors the two ends of the miRNA to orient and stabilize the miRNA into proper A-helix conformation. Second, the sub-seed bases, in particular positions 2–4, provide the initial searching platform that mediates the interaction with the target mRNA. Lastly, the hybridization between the mRNA and the sub-seed bases of the miRNA induces conformational change on Ago to allow further seed match for extended hybridization between the two RNAs. This increased complementarity significantly lengthens the residence time of Ago bound on the target mRNA, which may be necessary for sufficient gene silencing [78, 79].

The figure is adapted from [77] with Elsevier, Copyright 2017
Model summarizing the conformational changes of Ago during miRNA-target interaction.
5 Implications of the Sub-seed Region: 1-D Target Search
One of the unresolved questions in RISC-target interaction is how the miRNA embedded in RISC can effectively find its target mRNAs in a complex media like inside the cell. The sequential target recognition process by RISC suggests one clue: By using the sub-seed sequences, Ago may find an mRNA with the partial seed match first and then slide along the RNA to search for better binding sites, i.e., sites with extended seed match sequences if such sites do exist. By doing so, the target search in the three-dimensional space has effectively become one-dimensional sliding problem [79]. In fact, this kind of one-dimensional search algorithm is employed by transcription factors in search for their optimal binding sites on the DNA. Previous studies on the mechanism of the recognition of the lac operon by LacI repressor in Escherichia coli (E. coli) showed that the protein first finds the DNA in the three-dimensional space and slides along the DNA to find the optimal binding site located within the lac operon [83,84,85]. Therefore, the three-dimensional diffusion has essentially become one-dimensional sliding which significantly facilitates the target search process.
Using the combination of three- and one-dimensional search mechanisms can be advantageous in multiple ways. First, once the RISC finds an mRNA with sub-seed match, it can undergo fast lateral diffusion along the RNA to search for the optimal binding site with extended seed match (Fig. 8.7) [79]. This lateral searching process may require hopping and sliding along the mRNA rather than trying to find the optimal site through searching in three-dimensional space of the cytosol [79]. The facilitated search mechanism may allow RISC to act as a multi-turnover type of regulator as it can quickly move from one target to another. In addition, the ability to search in three-dimensional space is necessary as RISC, unlike transcription factors, targets mRNAs. As the sub-seed region only contains three or four nucleotides, it is likely that mRNAs with the sub-seed match may not have the sequences that are complementary to the rest of the seed region. In this case, the RISC should detach from the mRNA and diffuse through the cytosol, searching for an unprobed new target mRNA. Of note, the partial seed match assists this step as it has lower binding affinities to the mRNA compared to that of the full seed match [78, 79]. The search for the new target strictly depends on the diffusion in the three-dimensional space. Once the RISC finds another mRNA with sub-seed match, it will again undergo one-dimensional sliding and hopping along the RNA to find the optimal binding site (Fig. 8.7). Therefore, the sub-seed match allows the RISC to scan through many different mRNAs to find the true target, and the optimal target search will depend on the proper distribution of three-dimensional search and one-dimensional scan mode of the RISC [79].

The figure is adapted from [79] with Elsevier, Copyright 2015
Model summarizing dynamic target search by RISC using sub-seed match and lateral diffusion along the target mRNA.
The structure of Ago and its conformational changes during RISC-target recognition play a role in optimizing the balance between three-dimensional search and one-dimensional scan processes. Previous investigation of the one-dimensional scanning of transcription factor argued that it is not possible for the transcription factor to have both fast searching and stable binding [86,87,88]. This is because fast searching requires weak interaction between the protein and the DNA and consequently transcription factor with fast searching speed is likely to miss many of its true binding sites. Similarly, stable protein–DNA interaction implies slow dissociation which results in slow lateral diffusion as the protein gets trapped at nontarget sites. This results in speed–stability paradox in DNA scanning where fast and specific target search is difficult to achieve simultaneously [87, 89]. However, this problem can be resolved if the protein can adopt multiple configurations each with different DNA binding affinities. During the initial search mode, the protein may show weak interaction with the DNA and only when it recognizes sequences similar to its binding site, then the protein may change its configuration and slowly scan the vicinity to find the optimal binding site [87, 89] (Fig. 8.8).

The figure is modified from [89] with Elsevier, Copyright 2016
Speed–stability paradox of DNA–protein interaction. In order to overcome the speed–stability paradox, transcription factors often use two state DNA interactions. The search mode is characterized by weak DNA–protein interaction which allows fast search with relatively smooth energy landscape. The recognition mode shows increased interaction with the DNA which results in decreased speed with increased specificity. The recognition mode also shows a large energy variation.
The structure of Ago provides an ideal example for the configuration changes required for optimal target search process. When miRNA is loaded onto Ago, the protein arranges the miRNA such that only the mini-seed bases can adapt A-form helical structure [22]. In other words, the kink at the position 7 imposed by Ago prevents the target interaction beyond the fifth position of the guide RNA, restricting the interaction and lowering binding affinity between miRNA and mRNA pairs [75]. Therefore, during the scan mode, Ago effectively limits miRNA–mRNA interactions to reduce the binding affinity such that it can quickly scan through the 3′ UTR to find the sequences that match the sub-seed region of the miRNA [79].
In addition, once the sub-seed match region is found, the hybridization between miRNA and mRNA induces the conformational change in Ago such that it now allows full seed interaction [79]. This change in Ago configuration allows base pairing beyond the sub-seed region and can significantly increase the RISC–mRNA-binding affinity. As a result, the scanning process has slowed down sufficiently to find the full or nearly full seed match sites on the mRNA [79]. Therefore, the steric hindrance imposed by Ago and conformational change of Ago by miRNA–mRNA interaction provide the necessary conditions for the optimal target search by RISC as suggested by Slutsky and Mirny: facilitated diffusion and one-dimensional target search along the mRNA and slowed diffusion near the target site in order to converge on the site with full seed match nucleotides [86,87,88,89]. Overall, structural understanding of Ago and the change induced by sub-seed base pairing strongly suggest that the target search mechanism by the RISC is a multi-step process with at least one scan mode and one recognition mode.
6 Toward Target Cleavage
Gene silencing by RISC is mediated through at least three mechanisms: (1) RISC components interact with the cap-binding proteins and suppress translation at the initiation step; (2) RISC induces RNA decay by triggering deadenylation and decapping of the target mRNA; (3) RISC directly cleaves mRNA at the binding site. The direct cleavage requires hAgo2, the only cleavage competent Ago in the human genome [21, 37]. While the miRNA-mediated gene silencing mostly occurs through the first two mechanisms, small interfering RNAs (siRNAs) can also induce target cleavage when loaded onto hAgo2. The difference between the two pathways (miRNA vs. siRNA) lies in the extent of the target complementarity. Target cleavage can occur when the 10th and 11th nucleotides of the guide siRNA/miRNA pair with the mRNA [90,91,92]. However, while siRNAs are designed to have extended base pairing with the target, most of miRNAs do not base pair at these positions, resulting in only the siRNA being able to induce target cleavage.
Consistent with this idea, bioinformatics studies have shown that most of the targets of miRNAs in human do not show complementarity beyond the seed sequences and thus are not cleaved by hAgo2 [71,72,73,74]. Moreover, numerous studies have shown that seed sequences of the miRNA are the key determinant of target selection and the rest of the sequences do not affect RISC-target interactions [32, 72, 82, 93,94,95,96]. Yet, structural and single-molecule studies have shown that seed pairing triggers an additional conformational change in Ago, which provides a key understanding of the mechanism and efficiency of gene silencing by RISC [75, 81, 97, 98].
First, the sub-seed match relieves the kink at the position 7 and allows extended seed match with the mRNA [77]. However, the pairing beyond the eighth position is still restricted and requires the widening of the channel between the PAZ and the N-terminal domains [75]. Recently, Jo et al. observed that many of the targets of the miRNA are not cleaved despite the perfect complementarities [99, 100]. One possibility is that Ago imposes structural hindrance such that the 10th and 11th positions of the miRNA cannot base pair with the corresponding complementary nucleotides on the mRNA. This result suggests that the identity of miRNA may be important in predicting its target cleavage capability. Therefore, the simple identity and complementarity are not enough to predict the target cleavage and further investigation on the conformational change on Ago due to miRNA–mRNA interaction is required [77].
Although the seed sequence match may not induce conformational change in Ago to allow target cleavage, it does rearrange the protein such that the 13th–16th nucleotides of the miRNA (also known as the supplementary region) are now configured into an A-helical form and may base pair with the target RNA (Fig. 8.9) [75]. The extended target complementarity in this region of the miRNA further enhances the binding affinity and increases the residence time of Ago on the target mRNA [75, 78, 79].

The figure is adapted from [75] with American Association for the Advancement of Science, Copyright 2014
Seed match with the target mRNA triggers conformational change of Ago such that the supplementary region of the guide miRNA arranges into A-form helical structure and may base pair with the target mRNA.
The sequences beyond the 16th position of the miRNA cannot interact with mRNA due to structural constraint imposed by Ago protein [77]. Similar to the first sequence of the miRNA, the 3′ end of the miRNA is anchored at the PAZ domain [22]. Ago does not release the 3′ end of the miRNA even after the pairing at the supplementary region and prevents the access of the sequences beyond the 16th position [31, 45, 101, 102]. This tight association between the PAZ domain and the 3′ end of the miRNA is necessary to ensure efficient target release (Fig. 8.10). It has been shown that RNA duplex longer than 12 base pairs has half-life of approximately one year, indicating that the two will form an extremely stable complex and are not likely to dissociate [103]. However, miRNA loaded onto Ago dissociates with the target mRNA quickly and allows the protein to act as a multi-turnover enzyme [100]. This reversible interaction between Ago and its target might be possible because the 3′ end of the miRNA is anchored at the PAZ domain, which lowers the binding affinity between the Ago and the target mRNA [31, 45, 67, 101, 102, 104].

The figure is adapted from [31] with American Chemical Society, Copyright 2013
During miRNA-target interaction, the 3′ end of the miRNA is anchored at the PAZ domain and is prohibited from base pairing with the target, which may play a role in efficient target release.
Lastly, evidences suggest that Ago may directly interact with mRNAs and contribute to the target recognition process [79]. The first sequence of the miRNA is anchored and cannot interact with the target [21, 90, 105, 106]. However, when the first sequence of the miRNA is uridine, it may interact with adenine nucleotide of the mRNA and anchor the mRNA onto the MID domain of Ago [107]. This provides an additional sequence pairing between miRNA and mRNA and can increase the efficiency of gene silencing effect [108]. Moreover, this interaction may account for the phenomenon where uridine is the preferred sequence at the 5′ end of the guide strand [21, 90, 105, 106]. Interestingly, using a single-molecule approach, Schirle et al. showed that the adenine anchoring in Ago does not influence the initial target recognition process, but does increase the residence time of Ago on the mRNA [108]. Therefore, RISC can still search for its target using the sub-seed sequences, and the base pairing at the first position only affects the gene silencing efficiency.
7 Concluding Remarks
In this chapter, we present structural, biochemical, and biophysical single-molecule studies on the Ago–miRNA-target interactions. As a key posttranscriptional regulatory molecule, miRNA has received increasing attention over the recent decades [109]. In particular, miRNAs are recognized as key potential biomarkers for diagnosis of human disease and prognosis during clinical treatments as well as indicators of cellular status [109]. The dynamic changes in miRNA expressions are associated with a variety of human diseases including heart disease, neurological diseases, immune function disorders, and age-related diseases. Furthermore, dramatic changes in miRNA profiles have been observed during disease progression, drug treatment, and differentiation of stem cells.
Despite its significance, our understanding of miRNA-mediated gene silencing is limited due to lack of knowledge of the in-depth mechanism of RISC assembly and RISC-target interactions. Numerous structural studies have significantly expanded our understanding of how miRNAs are loaded onto Ago. Loading of the miRNA to the MID and PAZ domains of Ago positions the RNA such that it can efficiently search for targets. In addition, miRNA–Ago complex constantly undergoes changes in its configuration as it interacts with the target mRNA to effectively find and associate with the true targets. Lastly, anchoring of the 3′ end of the miRNA to the PAZ domain ensures the efficient target release, which may be important for the effective gene silencing by RISC.
These structural studies inspired the development of new models of target search and regulation that are further confirmed by single-molecule experiments. Particularly, these studies investigated the binding dynamics of miRNA-target interactions. Like transcription factors, RISC also utilizes one-dimensional scanning in order to quickly search for the optimal binding site on an mRNA. This mode of quick scanning is possible as Ago protein induces steric hindrance and prevents the complementary pairing beyond the sub-seed region. More importantly, RISC also can readily dissociate from the mRNA that lacks full seed match sequences and undergo diffusion in the three-dimensional space in the cytosol to find a new potential target. Ago structure also plays a key role in this switch between the two target search modes. First, complementary pairing at the sub-seed region triggers structural change such that the helix-7 motif can no longer block the miRNA–mRNA interaction beyond the sub-seed region. Furthermore, seed pairing also can switch the protein to the recognition mode and allow base pairing even at the supplementary region. Therefore, Ago subdivides miRNA into multiple functional domains and changes its configuration to determine which regions to expose to the media in order to optimize its target search and release processes (Fig. 8.11).

The figure is adapted from [77] with Elsevier, Copyright 2017
Schematic of miRNA-target interaction depicting subdivision of the miRNA by Ago.
The one-dimensional scanning mechanism as well as the dynamic conformational change induced by target interaction is also observed in various other systems. As discussed above, LacI repressor scans through the DNA to find the optimal binding site on the lac operon. In addition, the interaction between RecA and DNA is restricted to 7–8 nt due to the steric hindrance imposed by RecA protein [110]. Qi and colleagues termed these sequences as “microhomology motif” which serves as an initial platform for target recognition by RecA [111]. Such restriction may allow RecA to quickly scan the DNA to find the optimal binding site. Lastly, CRISPR/Cas protein also utilizes multi-step target recognition mechanism. It first scans the DNA to find the PAM motif and subsequently scans the vicinity to find the optimal binding site. Furthermore, binding to the extended complementary target induces conformational change to bring the nuclease domain to the target DNA [112,113,114]. These series of target recognition steps may account for the remarkable efficiency and specificity of the CRISPR system.
Given the generality of the target search mechanism, we expect that one-dimensional scanning as well as the conformational change induced by the target interaction is an important regulatory strategy in RNA/DNA binding proteins. Quantitative and single-molecule approaches in combination with the structural crystallographic information will provide valuable insights into a comprehensive understanding of RNA–protein interactions.
References
Bartel, D. P. (2004). MicroRNAs: Genomics, biogenesis, mechanism, and function. Cell, 116, 281–297.
Ha, M., & Kim, V. N. (2014). Regulation of microRNA biogenesis. Nature Reviews Molecular Cell Biology, 15, 509–524.
Cai, X., Hagedorn, C. H., & Cullen, B. R. (2004). Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA, 10, 1957–1966.
Lee, Y., Jeon, K., Lee, J. T., Kim, S., & Kim, V. N. (2002). MicroRNA maturation: Stepwise processing and subcellular localization. The EMBO Journal, 21, 4663–4670.
Lee, Y., et al. (2004). MicroRNA genes are transcribed by RNA polymerase II. The EMBO Journal, 23, 4051–4060.
Ozsolak, F., et al. (2008). Chromatin structure analyses identify miRNA promoters. Genes & Development, 22, 3172–3183.
Denli, A. M., Tops, B. B., Plasterk, R. H., Ketting, R. F., & Hannon, G. J. (2004). Processing of primary microRNAs by the microprocessor complex. Nature, 432, 231–235.
Gregory, R. I., et al. (2004). The Microprocessor complex mediates the genesis of microRNAs. Nature, 432, 235–240.
Han, J., et al. (2004). The Drosha-DGCR9 complex in primary microRNA processing. Genes & Development, 18, 3016–3027.
Landthaler, M., Yalcin, A., & Tuschl, T. (2004). The human DiGeorge syndrome critical region gene 8 and Its D. melanogaster homolog are required for miRNA biogenesis. Current Biology, 14, 2162–2167.
Lee, Y., et al. (2003). The nuclear RNase III Drosha initiates microRNA processing. Nature, 425, 415–419.
Kim, B., Jeong, K., & Kim, V. N. (2017). Genome-wide mapping of DROSHA cleavage sites on primary microRNAs and noncanonical substrates. Molecular Cell, 66, 258–269. e255.
Nguyen, T. A., et al. (2015). Functional anatomy of the human microprocessor. Cell, 161, 1374–1387.
Kwon, S. C., et al. (2016). Structure of human DROSHA. Cell, 164, 81–90.
Auyeung, V. C., Ulitsky, I., McGeary, S. E., & Bartel, D. P. (2013). Beyond secondary structure: Primary-sequence determinants license pri-miRNA hairpins for processing. Cell, 152, 844–858.
Yi, R., Qin, Y., Macara, I. G., & Cullen, B. R. (2003). Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes & Development, 17, 3011–3016.
MacRae, I. J., Zhou, K., & Doudna, J. A. (2007). Structural determinants of RNA recognition and cleavage by Dicer. Nature Structural & Molecular Biology, 14, 934–940.
Park, J. E., et al. (2011). Dicer recognizes the 5′ end of RNA for efficient and accurate processing. Nature, 475, 201–205.
Macrae, I. J., et al. (2006). Structural basis for double-stranded RNA processing by Dicer. Science, 311, 195–198.
Hutvagner, G., & Simard, M. J. (2008). Argonaute proteins: Key players in RNA silencing. Nature Reviews Molecular Cell Biology, 9, 22–32.
Liu, J., et al. (2004). Arogonaute2 Is the catalytic engine of mammalian RNAi. Science, 305, 1437–1441.
Schirle, N. T., & MacRae, I. J. (2012). The crystal structure of human Argonaute2. Science, 336, 1037–1040.
Song, J. J., Smith, S. K., Hannon, G. J., & Joshua-Tor, L. (2004). Crystal structure of Argonaute and its implications for RISC slicer activity. Science, 305, 1434–1437.
Gan, H. H., & Gunsalus, K. C. (2015). Assembly and analysis of eukaryotic Argonaute-RNA complexes in microRNA-target recognition. Nucleic Acids Research, 43, 9613–9625.
Lingel, A., Simon, B., Izaurralde, E., & Sattler, M. (2003). Structure and nucleic-acid binding of the Drosophila Argonaute 2 PAZ domain. Nature, 426, 465–469.
Lingel, A., Simon, B., Izaurralde, E., & Sattler, M. (2004). Nucleic acid 3′-end recognition by the Argonaute2 PAZ domain. Nature Structural & Molecular Biology, 11, 576–577.
Ma, J. B., Ye, K., & Patel, D. J. (2004). Structural basis for overhang-specific small interfering RNA recognition by the PAZ domain. Nature, 429, 318–322.
Ma, J. B., et al. (2005). Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature, 434, 666–670.
Parker, J. S., Roe, S. M., & Barford, D. (2004). Crystal structure of a PIWI protein suggests mechanisms for siRNA recognition and slicer activity. The EMBO Journal, 23, 4727–4737.
Yuan, Y. R., et al. (2005). Crystal structure of A. aeolicus argonaute, a site-specific DNA-guided endoribonuclease, provides insights into RISC-mediated mRNA cleavage. Molecular Cell, 19, 405–419.
Jung, S. R., et al. (2013). Dynamic anchoring of the 3′-end of the guide strand controls the target dissociation of Argonaute-guide complex. The Journal of the American Chemical Society, 135, 16865–16871.
Haley, B., & Zamore, P. D. (2004). Kinetic analysis of the RNAi enzyme complex. Nature Structural & Molecular Biology, 11, 599–606.
Zamore, P. D. (2001). Thirty-three years later, a glimpse at the ribonuclease III active site. Molecular Cell, 8, 1158–1160.
Elbashir, S. M. (2001). RNA interference is mediated by 21- and 22-nucleotide RNAs. Genes & Development, 15, 188–200.
Elbashir, S. M., Martinez, J., Patkaniowska, A., Lendeckel, W., & Tuschl, T. (2001). Functional anatomy of siRNAs for mediating efficient RNAi in Drosophila melanogaster embryo lysate. The EMBO Journal, 20, 6877–6888.
Nykanen, A., Haley, B., & Zamore, P. D. (2001). ATP requirements and small interfering RNA structure in the RNA interference pathway. Cell, 107, 309–321.
Meister, G., et al. (2004). Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Molecular Cell, 15, 185–197.
Nowotny, M., Gaidamakov, S. A., Crouch, R. J., & Yang, W. (2005). Crystal structures of RNase H bound to an RNA/DNA hybrid: Substrate specificity and metal-dependent catalysis. Cell, 121, 1005–1016.
Rivas, F. V., et al. (2005). Purified Argonaute2 and an siRNA form recombinant human RISC. Nature Structural & Molecular Biology, 12, 340–349.
Schwarz, D. S., Tomari, Y., & Zamore, P. D. (2004). The RNA-induced silencing complex is a Mg2+ -dependent endonuclease. Current Biology, 14, 787–791.
Martinez, J., & Tuschl, T. (2004). RISC is a 5′ phosphomonoester-producing RNA endonuclease. Genes & Development, 18, 975–980.
Jinek, M., & Doudna, J. A. (2009). A three-dimensional view of the molecular machinery of RNA interference. Nature, 457, 405–412.
Forstemann, K., Horwich, M. D., Wee, L., Tomari, Y., & Zamore, P. D. (2007). Drosophila microRNAs are sorted into functionally distinct argonaute complexes after production by dicer-1. Cell, 130, 287–297.
Park, M. S., et al. (2017). Human Argonaute3 has slicer activity. Nucleic Acids Research, 45, 11867–11877.
Wang, Y., Sheng, G., Juranek, S., Tuschl, T., & Patel, D. J. (2008). Structure of the guide-strand-containing argonaute silencing complex. Nature, 456, 209–213.
Kim, V. N. (2008). Sorting out small RNAs. Cell, 133, 25–26.
Kiriakidou, M., et al. (2007). An mRNA m7G cap binding-like motif within human Ago2 represses translation. Cell, 129, 1141–1151.
Frank, F., et al. (2011). Structural analysis of 5′-mRNA-cap interactions with the human AGO2 MID domain. EMBO Reports, 12, 415–420.
Behm-Ansmant, I., et al. (2006). mRNA degradation by miRNAs and GW182 requires both CCR49:NOT deadenylase and DCP1:DCP2 decapping complexes. Genes & Development, 20, 1885–1898.
Braun, J. E., Huntzinger, E., Fauser, M., & Izaurralde, E. (2011). GW182 proteins directly recruit cytoplasmic deadenylase complexes to miRNA targets. Molecular Cell, 44, 120–133.
Fabian, M. R., et al. (2011). miRNA-mediated deadenylation is orchestrated by GW182 through two conserved motifs that interact with CCR51-NOT. Nature Structural & Molecular Biology, 18, 1211–1217.
Lim, J., et al. (2014). Uridylation by TUT4 and TUT7 marks mRNA for degradation. Cell, 159, 1365–1376.
Doench, J. G., & Sharp, P. A. (2004). Specificity of microRNA target selection in translational repression. Genes & Development, 18, 504–511.
Lewis, B. P., Shih, I. H., Jones-Rhoades, M. W., Bartel, D. P., & Burge, C. B. (2003). Prediction of mammalian microRNA targets. Cell, 115, 787–798.
Stark, A., Brennecke, J., Russell, R. B., & Cohen, S. M. (2003). Identification of Drosophila microRNA targets. PLOS Biology, 1, E60.
Gregory, R. I., Chendrimada, T. P., Cooch, N., & Shiekhattar, R. (2005). Human RISC couples microRNA biogenesis and posttranscriptional gene silencing. Cell, 123, 631–640.
MacRae, I. J., Ma, E., Zhou, M., Robinson, C. V., & Doudna, J. A. (2008). In vitro reconstitution of the human RISC-loading complex. Proceedings of the National Academy of Sciences of the United States of America, 105, 512–517.
Maniataki, E., & Mourelatos, Z. (2005). A human, ATP-independent, RISC assembly machine fueled by pre-miRNA. Genes & Development, 19, 2979–2990.
Wang, H. W., et al. (2009). Structural insights into RNA processing by the human RISC-loading complex. Nature Structural & Molecular Biology, 16, 1148–1153.
Kanellopoulou, C., et al. (2005). Dicer-deficient mouse embryonic stem cells are defective in differentiation and centromeric silencing. Genes & Development, 19, 489–501.
Martinez, J., Patkaniowska, A., Urlaub, H., Luhrmann, R., & Tuschl, T. (2002). Single-stranded antisense siRNAs guide target RNA cleavage in RNAi. Cell, 110, 563–574.
Murchison, E. P., Partridge, J. F., Tam, O. H., Cheloufi, S., & Hannon, G. J. (2005). Characterization of Dicer-deficient murine embryonic stem cells. Proceedings of the National Academy of Sciences of the United States of America, 102, 12135–12140.
Ye, X., et al. (2011). Structure of C3PO and mechanism of human RISC activation. Nature Structural & Molecular Biology, 18, 650–657.
Betancur, J. G., & Tomari, Y. (2012). Dicer is dispensable for asymmetric RISC loading in mammals. RNA, 18, 24–30.
Cheloufi, S., Dos Santos, C. O., Chong, M. M., & Hannon, G. J. (2010). A dicer-independent miRNA biogenesis pathway that requires Ago catalysis. Nature, 465, 584–589.
Kim, Y., & Kim, V. N. (2012). MicroRNA factory: RISC assembly from precursor microRNAs. Molecular Cell, 46, 384–386.
Kawamata, T., & Tomari, Y. (2010). Making RISC. Trends in Biochemical Sciences, 35, 368–376.
Tomari, Y., Matranga, C., Haley, B., Martinez, N., & Zamore, P. D. (2004). A protein sensor for siRNA asymmetry. Science, 306, 1377–1380.
Eulalio, A., Huntzinger, E., & Izaurralde, E. (2008). Getting to the root of miRNA-mediated gene silencing. Cell, 132, 9–14.
Filipowicz, W., Bhattacharyya, S. N., & Sonenberg, N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: Are the answers in sight? Nature Reviews Genetics, 9, 102–114.
Baek, D., et al. (2008). The impact of microRNAs on protein output. Nature, 455, 64–71.
Brennecke, J., Stark, A., Russell, R. B., & Cohen, S. M. (2005). Principles of microRNA-target recognition. PLOS Biology, 3, e85.
Selbach, M., et al. (2008). Widespread changes in protein synthesis induced by microRNAs. Nature, 455, 58–63.
Kim, D., et al. (2016). General rules for functional microRNA targeting. Nature Genetics, 48, 1517–1526.
Schirle, N. T., Sheu-Gruttadauria, J., & MacRae, I. J. (2014). Structural basis for microRNA targeting. Science, 346, 608–613.
Song, J. J., et al. (2003). The crystal structure of the Argonaute2 PAZ domain reveals an RNA binding motif in RNAi effector complexes. Nature Structural Biology, 10, 1026–1032.
Klein, M., Chandradoss, S. D., Depken, M., & Joo, C. (2017). Why Argonaute is needed to make microRNA target search fast and reliable. Seminars in Cell and Developmental Biology, 65, 20–28.
Salomon, W. E., Jolly, S. M., Moore, M. J., Zamore, P. D., & Serebrov, V. (2015). Single-molecule imaging reveals that Argonaute reshapes the binding properties of its nucleic acid guides. Cell, 162, 84–95.
Chandradoss, S. D., Schirle, N. T., Szczepaniak, M., MacRae, I. J., & Joo, C. (2015). A dynamic search process underlies microRNA targeting. Cell, 162, 96–107.
Elkayam, E., et al. (2012). The structure of human argonaute-2 in complex with miR-20a. Cell, 150, 100–110.
Nakanishi, K., Weinberg, D. E., Bartel, D. P., & Patel, D. J. (2012). Structure of yeast Argonaute with guide RNA. Nature, 486, 368–374.
Lai, E. C. (2002). Micro RNAs are complementary to 3′ UTR sequence motifs that mediate negative post-transcriptional regulation. Nature Genetics, 30, 363–364.
Berg, O. G., Winter, R. B., & von Hippel, P. H. (1981). Diffusion-driven mechanisms of protein translocation on nucleic acids. Biochemistry, 20, 6929–6948.
Riggs, A. D., Bourgeois, S., & Cohn, M. (1970). The lac represspr-operator interaction III. Kinetic studies. The Journal of Molecular Biology, 53, 401–417.
von Hippel, P. H., & Berg, O. G. (1989). Facilitated target location in biological systems. The Journal of Biological Chemistry, 264, 675–678.
Mirny, L., et al. (2009). How a protein searches for its site on DNA: The mechanism of facilitated diffusion. Journal of Physics A: Mathematical and Theoretical, 42
Slutsky, M., & Mirny, L. A. (2004). Kinetics of protein-DNA interaction: Facilitated target location in sequence-dependent potential. The Biophysical Journal, 87, 4021–4035.
Gerland, U., Moroz, J. D., & Hwa, T. (2002). Physical constraints and functional characterists of transcription factor-DNA interaction. Proceedings of the National Academy of Sciences of the United States of America, 99, 12015–12020.
Kong, M., & Van Houten, B. (2017). Rad4 recognition-at-a-distance: Physical basis of conformation-specific anomalous diffusion of DNA repair proteins. Progress in Biophysics & Molecular Biology, 127, 93–104.
Chiu, Y.-L., & Rana, T. M. (2002). RNAi in human cells. Molecular Cell, 10, 549–561.
Doench, J. G., Petersen, C. P., & Sharp, P. A. (2003). siRNAs can function as miRNAs. Genes & Development, 17, 438–442.
Hutvagner, G., & Zamore, P. D. (2002). A microRNA in a multiple-turnover RNAi enzyme complex. Science, 297, 2056–2060.
Bartel, D. P. (2009). MicroRNAs: Target recognition and regulatory functions. Cell, 136, 215–233.
Krek, A., et al. (2005). Combinatorial microRNA target predictions. Nature Genetics, 37, 495–500.
Lewis, B. P., Burge, C. B., & Bartel, D. P. (2005). Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell, 120, 15–20.
Lim, L. P., et al. (2005). Microarray analysis shows that some microRNAs downregulate large numbers of target mRNAs. Nature, 433, 769–773.
Faehnle, C. R., Elkayam, E., Haase, A. D., Hannon, G. J., & Joshua-Tor, L. (2013). The making of a slicer: Activation of human Argonaute-1. Cell Reports, 3, 1901–1909.
Nakanishi, K., et al. (2013). Eukaryote-specific insertion elements control human ARGONAUTE slicer activity. Cell Reports, 3, 1893–1900.
Jo, M. H., et al. (2015). Human Argonaute 2 has diverse reaction pathways on target RNAs. Molecular Cell, 59, 117–124.
Wee, L. M., Flores-Jasso, C. F., Salomon, W. E., & Zamore, P. D. (2012). Argonaute divides its RNA guide into domains with distinct functions and RNA-binding properties. Cell, 151, 1055–1067.
Sasaki, H. M., & Tomari, Y. (2012). The true core of RNA silencing revealed. Nature Structural & Molecular Biology, 19, 657–660.
Zander, A., Holzmeister, P., Klose, D., Tinnefeld, P., & Grohmann, D. (2014). Single-molecule FRET supports the two-state model of Argonaute action. RNA Biology, 11, 45–56.
Herschlag, D. (1991). Implications of ribozyme kinetics for targeting the cleavage of specific RNA molecules in vivo: More isn’t always better. Proceedings of the National Academy of Sciences of the United States of America, 88, 6921–6925.
Deerberg, A., Willkomm, S., & Restle, T. (2013). Minimal mechanistic model of siRNA-dependent target RNA slicing by recombinant human Argonaute 2 protein. Proceedings of the National Academy of Sciences of the United States of America, 110, 17850–17855.
Bofill-De Ros, X., & Gu, S. (2016). Guidelines for the optimal design of miRNA-based shRNAs. Methods, 103, 157–166.
Seitz, H., Tushir, J. S., & Zamore, P. D. (2011). A 5′-uridine amplifies miRNA/miRNA* asymmetry in Drosophila by promoting RNA-induced silencing complex formation. Silence, 2, 4.
Mi, S., et al. (2008). Sorting of small RNAs into Arabidopsis argonaute complexes is directed by the 5′ terminal nucleotide. Cell, 133, 116–127.
Schirle, N. T., Sheu-Gruttadauria, J., Chandradoss, S. D., Joo, C., & MacRae, I. J. (2015). Water-mediated recognition of t1-adenosine anchors Argonaute2 to microRNA targets. Elife, 4
Casey, M. C., Kerin, M. J., Brown, J. A., & Sweeney, K. J. (2015). Evolution of a research field-a micro (RNA) example. PeerJ, 3, e829.
Ragunathan, K., Liu, C., & Ha, T. (2012). RecA filament sliding on DNA facilitates homology search. Elife, 1, e00067.
Qi, Z., et al. (2015). DNA sequence alignment by microhomology sampling during homologous recombination. Cell, 160, 856–869.
Shvets, A. A., & Kolomeisky, A. B. (2017). Mechanism of genome interrogation: How CRISPR RNA-guided Cas9 proteins locate specific targets on DNA. The Biophysical Journal, 113, 1416–1424.
Sternberg, S. H., LaFrance, B., Kaplan, M., & Doudna, J. A. (2015). Conformational control of DNA target cleavage by CRISPR-Cas9. Nature, 527, 110–113.
Westra, E. R., et al. (2013). Type I-E CRISPR-cas systems discriminate target from non-target DNA through base pairing-independent PAM recognition. PLOS Genetics, 9, e1003742.
Acknowledgements
This work was supported by Samsung Research Funding & Incubation Center of Samsung Electronics under Project Number SRFC-MA1702-08.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this chapter
Cite this chapter
Kim, S., Kim, Y. (2019). Biophysical and Biochemical Approaches in the Analysis of Argonaute–MicroRNA Complexes. In: Joo, C., Rueda, D. (eds) Biophysics of RNA-Protein Interactions. Biological and Medical Physics, Biomedical Engineering. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-9726-8_8
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9726-8_8
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-9724-4
Online ISBN: 978-1-4939-9726-8
eBook Packages: Physics and AstronomyPhysics and Astronomy (R0)