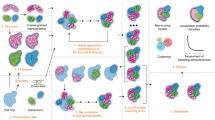
Abstract
Integrative structure modeling provides 3D models of macromolecular systems that are based on information from multiple types of experiments, physical principles, statistical inferences, and prior structural models. Here, we provide a hands-on realistic example of integrative structure modeling of the quaternary structure of the actin, tropomyosin, and gelsolin protein assembly based on electron microscopy, solution X-ray scattering, and chemical crosslinking data for the complex as well as excluded volume, sequence connectivity, and rigid atomic X-ray structures of the individual subunits. We follow the general four-stage process for integrative modeling, including gathering the input information, converting the input information into a representation of the system and a scoring function, sampling alternative model configurations guided by the scoring function, and analyzing the results. The computational aspects of this approach are implemented in our open-source Integrative Modeling Platform (IMP), a comprehensive and extensible software package for integrative modeling (https://integrativemodeling.org). In particular, we rely on the Python Modeling Interface (PMI) module of IMP that provides facile mixing and matching of macromolecular representations, restraints based on different types of information, sampling algorithms, and analysis including validations of the input data and output models. Finally, we also outline how to deposit an integrative structure and corresponding experimental data into PDB-Dev, the nascent worldwide Protein Data Bank (wwPDB) resource for archiving and disseminating integrative structures (https://pdb-dev.wwpdb.org). The example application provides a starting point for a user interested in using IMP for integrative modeling of other biomolecular systems.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Mitra K, Frank J (2006) RIBOSOME DYNAMICS: insights from atomic structure modeling into cryo-electron microscopy maps. Annu Rev Biophys Biomol Struct 35:299–317. https://doi.org/10.1146/annurev.biophys.35.040405.101950
Robinson CV, Sali A, Baumeister W (2007) The molecular sociology of the cell. Nature 450:973–982
Sali A, Glaeser R, Earnest T, Baumeister W (2003) From words to literature in structural proteomics. Nature 422:216–225
Schmeing TM, Ramakrishnan V (2009) What recent ribosome structures have revealed about the mechanism of translation. Nature 461:1234–1242. https://doi.org/10.1038/nature08403
Blundell TL, Johnson LN (1976) Protein crystallography. Academic Press, New York
Chiu W, Baker ML, Jiang W et al (2005) Electron cryomicroscopy of biological machines at subnanometer resolution. Structure 13:363–372. https://doi.org/10.1016/j.str.2004.12.016
Lučić V, Leis A, Baumeister W (2008) Cryo-electron tomography of cells: connecting structure and function. Histochem Cell Biol 130:185–196. https://doi.org/10.1007/s00418-008-0459-y
Stahlberg H, Walz T (2008) Molecular electron microscopy: state of the art and current challenges. ACS Chem Biol 3:268–281. https://doi.org/10.1021/cb800037d
Parrish JR, Gulyas KD, Finley RL (2006) Yeast two-hybrid contributions to interactome mapping. Curr Opin Biotechnol 17:387–393. https://doi.org/10.1016/j.copbio.2006.06.006
Fernandez-Martinez J, Phillips J, Sekedat MD et al (2012) Structure–function mapping of a heptameric module in the nuclear pore complex. J Cell Biol 196:419–434. https://doi.org/10.1083/jcb.201109008
Gingras A-C, Gstaiger M, Raught B, Aebersold R (2007) Analysis of protein complexes using mass spectrometry. Nat Rev Mol Cell Biol 8:645–654. https://doi.org/10.1038/nrm2208
Ward AB, Sali A, Wilson IA (2013) Integrative structural biology. Science 339:913–915. https://doi.org/10.1126/science.1228565
Lasker K, Förster F, Bohn S et al (2012) Molecular architecture of the 26S proteasome holocomplex determined by an integrative approach. Proc Natl Acad Sci U S A 109:1380–1387
Simon B, Madl T, Mackereth CD et al (2010) An efficient protocol for NMR-spectroscopy-based structure determination of protein complexes in solution. Angew Chem Int Ed Engl 49:1967–1970. https://doi.org/10.1002/anie.200906147
Baù D, Sanyal A, Lajoie BR et al (2011) The three-dimensional folding of the α-globin gene domain reveals formation of chromatin globules. Nat Struct Mol Biol 18:107–114. https://doi.org/10.1038/nsmb.1936
Viswanath S, Bonomi M, Kim SJ et al (2017) The molecular architecture of the yeast spindle pole body core determined by Bayesian integrative modeling. Mol Biol Cell 28:3298–3314. https://doi.org/10.1091/mbc.E17-06-0397
Kim SJ, Fernandez-Martinez J, Nudelman I et al (2018) Integrative structure and functional anatomy of a nuclear pore complex. Nature 555:475–482. https://doi.org/10.1038/nature26003
Molnar K, Bonomi M, Pellarin R et al (2014) Cys-scanning disulfide crosslinking and Bayesian modeling probe the transmembrane signaling mechanism of the histidine kinase, PhoQ. Structure 22:1239–1251
Webb B, Viswanath S, Bonomi M et al (2018) Integrative structure modeling with the integrative modeling platform: integrative structure modeling with IMP. Protein Sci 27:245–258. https://doi.org/10.1002/pro.3311
Shen M, Sali A (2006) Statistical potential for assessment and prediction of protein structures. Protein Sci 15:2507–2524. https://doi.org/10.1110/ps.062416606
Sippl MJ (1990) Calculation of conformational ensembles from potentials of mena force. J Mol Biol 213:859–883. https://doi.org/10.1016/S0022-2836(05)80269-4
Brooks BR, Brooks CL, Mackerell AD et al (2009) CHARMM: the biomolecular simulation program. J Comput Chem 30:1545–1614. https://doi.org/10.1002/jcc.21287
Weiner SJ, Kollman PA, Case DA et al (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106:765–784. https://doi.org/10.1021/ja00315a051
Šali A, Blundell TL (1993) Comparative protein modelling by satisfaction of spatial restraints. J Mol Biol 234:779–815. https://doi.org/10.1006/jmbi.1993.1626
Kelley LA, Mezulis S, Yates CM et al (2015) The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc 10:845–858. https://doi.org/10.1038/nprot.2015.053
Hanot S, Bonomi M, Greenberg CH et al (2018) Bayesian multi-scale modeling of macromolecular structures based on cryo-electron microscopy density maps. https://doi.org/10.1101/113951
Kawabata T (2008) Multiple subunit fitting into a low-resolution density map of a macromolecular complex using a Gaussian mixture model. Biophys J 95:4643–4658. https://doi.org/10.1529/biophysj.108.137125
Metropolis N, Rosenbluth AW, Rosenbluth MN et al (1953) Equation of state calculations by fast computing machines. J Chem Phys 21:1087–1092. https://doi.org/10.1063/1.1699114
Swendsen RH, Wang J-S (1986) Replica Monte Carlo simulation of spin-glasses. Phys Rev Lett 57:2607–2609. https://doi.org/10.1103/PhysRevLett.57.2607
Viswanath S, Chemmama IE, Cimermancic P, Sali A (2017) Assessing exhaustiveness of stochastic sampling for integrative modeling of macromolecular structures. Biophys J 113:2344–2353. https://doi.org/10.1016/j.bpj.2017.10.005
Alber F, Dokudovskaya S, Veenhoff LM et al (2007) The molecular architecture of the nuclear pore complex. Nature 450:695–701
Sali A, Berman HM, Schwede T et al (2015) Outcome of the first wwPDB hybrid/integrative methods task force workshop. Structure 23:1156–1167. https://doi.org/10.1016/j.str.2015.05.013
Burley SK, Kurisu G, Markley JL et al (2017) PDB-Dev: a prototype system for depositing integrative/hybrid structural models. Structure 25:1317–1318. https://doi.org/10.1016/j.str.2017.08.001
Vallat B, Webb B, Westbrook JD et al (2018) Development of a prototype system for archiving integrative/hybrid structure models of biological macromolecules. Structure. https://doi.org/10.1016/j.str.2018.03.011
Gil VA, Guallar V (2013) pyRMSD: a Python package for efficient pairwise RMSD matrix calculation and handling. Bioinformatics 29:2363–2364. https://doi.org/10.1093/bioinformatics/btt402
Rao JN, Dominguez R (2014) Complex of ATP-actin with the C-terminal actin-binding domain of tropomodulin. https://doi.org/10.2210/pdb4pki/pdb
Schneidman-Duhovny D, Hammel M, Sali A (2010) FoXS: a web server for rapid computation and fitting of SAXS profiles. Nucleic Acids Res 38:541–544
Robinson P, Trnka M, Pellarin R et al (2015) Molecular architecture of the yeast mediator complex. Elife. https://doi.org/10.7554/eLife.08719
Goddard TD, Huang CC, Meng EC et al (2018) UCSF ChimeraX: meeting modern challenges in visualization and analysis: UCSF ChimeraX visualization system. Protein Sci 27:14–25. https://doi.org/10.1002/pro.3235
Carter L, Kim SJ, Schneidman-Duhovny D et al (2015) Prion protein-antibody complexes characterized by chromatography-coupled small-angle X-ray scattering. Biophys J 109:793–805
Lasker K, Topf M, Sali A, Wolfson HJ (2009) Inferential optimization for simultaneous fitting of multiple components into a cryoEM map of their assembly. J Mol Biol 388:180–194
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2019 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Saltzberg, D. et al. (2019). Modeling Biological Complexes Using Integrative Modeling Platform. In: Bonomi, M., Camilloni, C. (eds) Biomolecular Simulations. Methods in Molecular Biology, vol 2022. Humana, New York, NY. https://doi.org/10.1007/978-1-4939-9608-7_15
Download citation
DOI: https://doi.org/10.1007/978-1-4939-9608-7_15
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-4939-9607-0
Online ISBN: 978-1-4939-9608-7
eBook Packages: Springer Protocols