Abstract
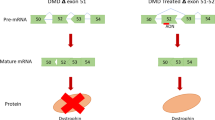
Antisense-mediated exon skipping and exon inclusion have proven to be powerful tools for treating neuromuscular diseases. The approval of Exondys 51 (eteplirsen) and Spinraza (nusinersen) for the treatment of patients with Duchenne muscular dystrophy (DMD) and spinal muscular atrophy (SMA) was the most noteworthy accomplishment in 2016. Exon skipping uses short DNA-like molecules called antisense oligonucleotides (AONs) to correct the disrupted reading frame, allowing the production of functional quasi-dystrophin proteins, and ameliorate the progression of the disease. Exon inclusion for SMA employs an AON targeting an intronic splice silencer site to include an exon which is otherwise spliced out. Recently, these strategies have also been explored in many other genetic disorders, including dysferlin-deficient muscular dystrophy (e.g., Miyoshi myopathy; MM, limb-girdle muscular dystrophy type 2B; LGMD2B, and distal myopathy with anterior tibial onset; DMAT), laminin α2 chain (merosin)-deficient congenital muscular dystrophy (MDC1A), sarcoglycanopathy (e.g., limb-girdle muscular dystrophy type 2C; LGMD2C), and Fukuyama congenital muscular dystrophy (FCMD). A major challenge in exon skipping and exon inclusion is the difficulty in designing effective AONs. The mechanism of mRNA splicing is highly complex, and the efficacy of AONs is often unpredictable. We will discuss the design of effective AONs for exon skipping and exon inclusion in this chapter.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Lee JJ, Yokota T (2013) Antisense therapy in neurology. J Pers Med 3(3):144–176. https://doi.org/10.3390/jpm3030144
Guncay A, Yokota T (2015) Antisense oligonucleotide drugs for Duchenne muscular dystrophy: how far have we come and what does the future hold? Future Med Chem 7(13):1631–1635. https://doi.org/10.4155/fmc.15.116
Vickers TA, Zhang H, Graham MJ et al (2006) Modification of MyD88 mRNA splicing and inhibition of IL-1beta signaling in cell culture and in mice with a 2'-O-methoxyethyl-modified oligonucleotide. J Immunol 176(6):3652–3661
(2016) Eteplirsen (Exondys 51) for Duchenne muscular dystrophy. Med Lett Drugs Ther 58 (1507):145–146
Shorrock HK, Gillingwater TH, Groen EJN. (2018) Overview of Current Drugs and Molecules in Development for Spinal Muscular Atrophy Therapy. Drugs 78(3):293–305
Nakamura A, Fueki N, Shiba N et al (2016) Deletion of exons 3-9 encompassing a mutational hot spot in the DMD gene presents an asymptomatic phenotype, indicating a target region for multiexon skipping therapy. J Hum Genet 61(7):663–667. https://doi.org/10.1038/jhg.2016.28
Koenig M, Beggs AH, Moyer M et al (1989) The molecular basis for Duchenne versus Becker muscular dystrophy: correlation of severity with type of deletion. Am J Hum Genet 45(4):498–506
Nakamura A, Shiba N, Miyazaki D et al (2017) Comparison of the phenotypes of patients harboring in-frame deletions starting at exon 45 in the Duchenne muscular dystrophy gene indicates potential for the development of exon skipping therapy. J Hum Genet 62(4):459–463. https://doi.org/10.1038/jhg.2016.152
Echigoya Y, Nakamura A, Nagata T et al (2017) Effects of systemic multiexon skipping with peptide-conjugated morpholinos in the heart of a dog model of Duchenne muscular dystrophy. Proc Natl Acad Sci U S A 114(16):4213–4218. https://doi.org/10.1073/pnas.1613203114
Shimo T, Tachibana K, Saito K et al (2014) Design and evaluation of locked nucleic acid-based splice-switching oligonucleotides in vitro. Nucleic Acids Res 42(12):8174–8187. https://doi.org/10.1093/nar/gku512
Rodrigues M, Echigoya Y, Fukada S et al (2016) Current translational research and murine models for Duchenne muscular dystrophy. J Neuromuscul Dis 3(1):29–48
Yokota T, Duddy W, Echigoya Y et al (2012) Exon skipping for nonsense mutations in Duchenne muscular dystrophy: too many mutations, too few patients? Expert Opin Biol Ther 12(9):1141–1152. https://doi.org/10.1517/14712598.2012.693469
Wein N, Vulin A, Findlay AR et al (2017) Efficient skipping of single exon duplications in DMD patient-derived cell lines using an antisense oligonucleotide approach. J Neuromuscul Dis 4(3):199–207. https://doi.org/10.3233/JND-170233
Yu X, Bao B, Echigoya Y et al (2015) Dystrophin-deficient large animal models: translational research and exon skipping. Am J Transl Res 7(8):1314–1331
Maruyama R, Echigoya Y, Caluseriu O et al (2017) Systemic delivery of morpholinos to skip multiple exons in a dog model of Duchenne muscular dystrophy. Methods Mol Biol 1565:201–213. https://doi.org/10.1007/978-1-4939-6817-6_17
Aartsma-Rus A, Fokkema I, Verschuuren J et al (2009) Theoretic applicability of antisense-mediated exon skipping for Duchenne muscular dystrophy mutations. Hum Mutat 30(3):293–299. https://doi.org/10.1002/humu.20918
Echigoya Y, Aoki Y, Miskew B et al (2015) Long-term efficacy of systemic multiexon skipping targeting dystrophin exons 45-55 with a cocktail of vivo-morpholinos in mdx52 mice. Mol Ther Nucleic Acids 4:e225. https://doi.org/10.1038/mtna.2014.76
Goyenvalle A, Griffith G, Babbs A et al (2015) Functional correction in mouse models of muscular dystrophy using exon-skipping tricyclo-DNA oligomers. Nat Med 21(3):270–275. https://doi.org/10.1038/nm.3765
Takagi M, Yagi M, Ishibashi K et al (2004) Design of 2'-O-Me RNA/ENA chimera oligonucleotides to induce exon skipping in dystrophin pre-mRNA. Nucleic Acids Symp Ser (Oxf) 48:297–298. https://doi.org/10.1093/nass/48.1.297
Miskew Nichols B, Aoki Y, Kuraoka M et al (2016) Multi-exon skipping using cocktail antisense oligonucleotides in the canine x-linked muscular dystrophy. J Vis Exp (111). doi: https://doi.org/10.3791/53776
Yokota T, Lu QL, Partridge T et al (2009) Efficacy of systemic morpholino exon-skipping in Duchenne dystrophy dogs. Ann Neurol 65(6):667–676. https://doi.org/10.1002/ana.21627
Alter J, Lou F, Rabinowitz A et al (2006) Systemic delivery of morpholino oligonucleotide restores dystrophin expression bodywide and improves dystrophic pathology. Nat Med 12(2):175–7
Yokota T, Nakamura A, Nagata T et al (2012) Extensive and prolonged restoration of dystrophin expression with vivo-morpholino-mediated multiple exon skipping in dystrophic dogs. Nucleic Acid Ther 22(5):306–315. https://doi.org/10.1089/nat.2012.0368
Maruyama R, Echigoya Y, Nakamura A et al (2017) Systemic injections of peptide-conjugated morpholinos improve cardiac symptoms of a dog model of Duchenne muscular dystrophy. Paper presented at the molecular therapy
Aoki Y, Nagata T, Yokota T et al (2013) Highly efficient in vivo delivery of PMO into regenerating myotubes and rescue in laminin-alpha2 chain-null congenital muscular dystrophy mice. Hum Mol Genet 22(24):4914–4928. https://doi.org/10.1093/hmg/ddt341
Wein N, Avril A, Bartoli M et al (2010) Efficient bypass of mutations in dysferlin deficient patient cells by antisense-induced exon skipping. Hum Mutat 31(2):136–142. https://doi.org/10.1002/humu.21160
Touznik A, Lee JJ, Yokota T (2014) New developments in exon skipping and splice modulation therapies for neuromuscular diseases. Expert Opin Biol Ther 14(6):809–819. https://doi.org/10.1517/14712598.2014.896335
Spraggon L, Cartegni L (2013) Antisense modulation of RNA processing as a therapeutic approach in cancer therapy. Drug Discov Today Ther Strateg 10(3):e139–e148. https://doi.org/10.1016/j.ddstr.2013.06.002
Zammarchi F, de Stanchina E, Bournazou E et al (2011) Antitumorigenic potential of STAT3 alternative splicing modulation. Proc Natl Acad Sci U S A 108(43):17779–17784. https://doi.org/10.1073/pnas.1108482108
Lentz JJ, Jodelka FM, Hinrich AJ et al (2013) Rescue of hearing and vestibular function by antisense oligonucleotides in a mouse model of human deafness. Nat Med 19(3):345–350. https://doi.org/10.1038/nm.3106
Wojtkowiak-Szlachcic A, Taylor K, Stepniak-Konieczna E et al (2015) Short antisense-locked nucleic acids (all-LNAs) correct alternative splicing abnormalities in myotonic dystrophy. Nucleic Acids Res 43(6):3318–3331. https://doi.org/10.1093/nar/gkv163
Gao QQ, Wyatt E, Goldstein JA et al (2015) Reengineering a transmembrane protein to treat muscular dystrophy using exon skipping. J Clin Invest 125(11):4186–4195. https://doi.org/10.1172/JCI82768
Touznik A, Maruyama R, Hosoki K et al (2017) LNA/DNA mixmer-based antisense oligonucleotides correct alternative splicing of the SMN2 gene and restore SMN protein expression in type 1 SMA fibroblasts. Sci Rep 7(1):3672. https://doi.org/10.1038/s41598-017-03850-2
Skordis LA, Dunckley MG, Yue B et al (2003) Bifunctional antisense oligonucleotides provide a trans-acting splicing enhancer that stimulates SMN2 gene expression in patient fibroblasts. Proc Natl Acad Sci U S A 100(7):4114–4119. https://doi.org/10.1073/pnas.0633863100
Madocsai C, Lim SR, Geib T et al (2005) Correction of SMN2 Pre-mRNA splicing by antisense U7 small nuclear RNAs. Mol Ther 12(6):1013–1022. https://doi.org/10.1016/j.ymthe.2005.08.022
Hua Y, Vickers TA, Baker BF et al (2007) Enhancement of SMN2 exon 7 inclusion by antisense oligonucleotides targeting the exon. PLoS Biol 5(4):e73
Baughan TD, Dickson A, Osman EY et al (2009) Delivery of bifunctional RNAs that target an intronic repressor and increase SMN levels in an animal model of spinal muscular atrophy. Hum Mol Genet 18(9):1600–1611. https://doi.org/10.1093/hmg/ddp076
Williams JH, Schray RC, Patterson CA et al (2009) Oligonucleotide-mediated survival of motor neuron protein expression in CNS improves phenotype in a mouse model of spinal muscular atrophy. J Neurosci 29(24):7633–7638. https://doi.org/10.1523/JNEUROSCI.0950-09.2009
Hua Y, Sahashi K, Hung G et al (2010) Antisense correction of SMN2 splicing in the CNS rescues necrosis in a type III SMA mouse model. Genes Dev 24(15):1634–1644. https://doi.org/10.1101/gad.1941310
Passini MA, Bu J, Richards AM et al (2011) Antisense oligonucleotides delivered to the mouse CNS ameliorate symptoms of severe spinal muscular atrophy. Sci Transl Med 3(72):72ra18. https://doi.org/10.1126/scitranslmed.3001777
Mitrpant C, Porensky P, Zhou H et al (2013) Improved antisense oligonucleotide design to suppress aberrant SMN2 gene transcript processing: towards a treatment for spinal muscular atrophy. PLoS One 8(4):e62114. https://doi.org/10.1371/journal.pone.0062114
Taniguchi-Ikeda M, Kobayashi K, Kanagawa M et al (2011) Pathogenic exon-trapping by SVA retrotransposon and rescue in Fukuyama muscular dystrophy. Nature 478(7367):127–131. https://doi.org/10.1038/nature10456
Lee JJA, Yokota T (2016) Translational research in nucleic acid therapies for muscular dystrophies. In: Takeda S'i, Miyagoe-Suzuki Y, Mori-Yoshimura M (eds) Translational research in muscular dystrophy, 1st edn. Springer, Tokyo, pp 87–102. https://doi.org/10.1007/978-4-431-55678-7
Dominov JA, Uyan O, Sapp PC et al (2014) A novel dysferlin mutant pseudoexon bypassed with antisense oligonucleotides. Ann Clin Transl Neurol 1(9):703–720. https://doi.org/10.1002/acn3.96
Lim KR, Maruyama R, Yokota T (2017) Eteplirsen in the treatment of Duchenne muscular dystrophy. Drug Des Devel Ther 11:533–545. https://doi.org/10.2147/DDDT.S97635
Nguyen Q, Yokota T (2017) Immortalized muscle cell model to test the exon skipping efficacy for Duchenne muscular dystrophy. J Pers Med 7(4). https://doi.org/10.3390/jpm7040013
Summerton J, Weller D (1997) Morpholino antisense oligomers: design, preparation, and properties. Antisense Nucleic Acid Drug Dev 7(3):187–195
Lu QL, Rabinowitz A, Chen YC et al (2005) Systemic delivery of antisense oligoribonucleotide restores dystrophin expression in body-wide skeletal muscles. Proc Natl Acad Sci U S A 102(1):198–203. https://doi.org/10.1073/pnas.0406700102
Chiriboga CA, Swoboda KJ, Darras BT et al (2016) Results from a phase 1 study of nusinersen (ISIS-SMNRx) in children with spinal muscular atrophy. Neurology 86(10):890–897. https://doi.org/10.1212/WNL.0000000000002445
Echigoya Y, Lim KRQ, Trieu N et al (2017) Quantitative antisense screening and optimization for exon 51 skipping in Duchenne muscular dystrophy. Mol Ther. https://doi.org/10.1016/j.ymthe.2017.07.014
Yang CJ, Wang L, Wu Y et al (2007) Synthesis and investigation of deoxyribonucleic acid/locked nucleic acid chimeric molecular beacons. Nucleic Acids Res 35(12):4030–4041. https://doi.org/10.1093/nar/gkm358
Echigoya Y, Mouly V, Garcia L et al (2015) In silico screening based on predictive algorithms as a design tool for exon skipping oligonucleotides in Duchenne muscular dystrophy. PLoS One 10(3):e0120058. https://doi.org/10.1371/journal.pone.0120058
Aartsma-Rus A, van Vliet L, Hirschi M et al (2009) Guidelines for antisense oligonucleotide design and insight into splice-modulating mechanisms. Mol Ther 17(3):548–553. https://doi.org/10.1038/mt.2008.205
Yokota T, Takeda S, Lu QL et al (2009) A renaissance for antisense oligonucleotide drugs in neurology: exon skipping breaks new ground. Arch Neurol 66(1):32–38
Saito T, Nakamura A, Aoki Y et al (2010) Antisense PMO found in dystrophic dog model was effective in cells from exon 7-deleted DMD patient. PLoS One 5(8):e12239. https://doi.org/10.1371/journal.pone.0012239
Echigoya Y, Yokota T (2014) Skipping multiple exons of dystrophin transcripts using cocktail antisense oligonucleotides. Nucleic Acid Ther 24(1):57–68. https://doi.org/10.1089/nat.2013.0451
Aoki Y, Yokota T, Wood MJ (2013) Development of multiexon skipping antisense oligonucleotide therapy for Duchenne muscular dystrophy. Biomed Res Int 2013:402369. https://doi.org/10.1155/2013/402369
Aslesh T, Maruyama R, Yokota T (2018) Skipping multiple exons to treat DMD—promises and challenges. Biomedicine 6(1):1
Aoki Y, Yokota T, Nagata T et al (2012) Bodywide skipping of exons 45-55 in dystrophic mdx52 mice by systemic antisense delivery. Proc Natl Acad Sci U S A 109(34):13763–13768. https://doi.org/10.1073/pnas.1204638109
Aartsma-Rus A, Van Deutekom JC, Fokkema IF et al (2006) Entries in the Leiden Duchenne muscular dystrophy mutation database: an overview of mutation types and paradoxical cases that confirm the reading-frame rule. Muscle Nerve 34(2):135–144
Desmet FO, Hamroun D, Lalande M et al (2009) Human Splicing Finder: an online bioinformatics tool to predict splicing signals. Nucleic Acids Res 37(9):e67. https://doi.org/10.1093/nar/gkp215
Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31(13):3406–3415
Kibbe WA (2007) OligoCalc: an online oligonucleotide properties calculator. Nucleic Acids Res 35(Web Server):W43–W46. https://doi.org/10.1093/nar/gkm234
Hubbard T, Barker D, Birney E et al (2002) The Ensembl genome database project. Nucleic Acids Res 30(1):38–41
Speir ML, Zweig AS, Rosenbloom KR et al (2016) The UCSC Genome Browser database: 2016 update. Nucleic Acids Res 44(D1):D717–D725. https://doi.org/10.1093/nar/gkv1275
Yokota T, Pistilli E, Duddy W et al (2007) Potential of oligonucleotide-mediated exon-skipping therapy for Duchenne muscular dystrophy. Expert Opin Biol Ther 7(6):831–842. https://doi.org/10.1517/14712598.7.6.831
Yokota T, Duddy W, Partridge T (2007) Optimizing exon skipping therapies for DMD. Acta Myol 26(3):179–184
Cartegni L, Wang J, Zhu Z et al (2003) ESEfinder: a web resource to identify exonic splicing enhancers. Nucleic Acids Res 31(13):3568–3571
Fairbrother WG, Yeo GW, Yeh R et al (2004) RESCUE-ESE identifies candidate exonic splicing enhancers in vertebrate exons. Nucleic Acids Res 32(Web Server):W187–W190. https://doi.org/10.1093/nar/gkh393
Yokota T, Hoffman E, Takeda S (2011) Antisense oligo-mediated multiple exon skipping in a dog model of Duchenne muscular dystrophy. Methods Mol Biol 709:299–312. https://doi.org/10.1007/978-1-61737-982-6_20
Flanigan KM, Voit T, Rosales XQ et al (2014) Pharmacokinetics and safety of single doses of drisapersen in non-ambulant subjects with Duchenne muscular dystrophy: results of a double-blind randomized clinical trial. Neuromuscul Disord 24(1):16–24. https://doi.org/10.1016/j.nmd.2013.09.004
Voit T, Topaloglu H, Straub V et al (2014) Safety and efficacy of drisapersen for the treatment of Duchenne muscular dystrophy (DEMAND II): an exploratory, randomised, placebo-controlled phase 2 study. Lancet Neurol 13(10):987–996. https://doi.org/10.1016/S1474-4422(14)70195-4
Mendell JR, Rodino-Klapac LR, Sahenk Z et al (2013) Eteplirsen for the treatment of Duchenne muscular dystrophy. Ann Neurol 74(5):637–647. https://doi.org/10.1002/ana.23982
Harding PL, Fall AM, Honeyman K et al (2007) The influence of antisense oligonucleotide length on dystrophin exon skipping. Mol Ther 15(1):157–166. https://doi.org/10.1038/sj.mt.6300006
Chan CY, Carmack CS, Long DD et al (2009) A structural interpretation of the effect of GC-content on efficiency of RNA interference. BMC Bioinformatics 10(Suppl 1):S33. https://doi.org/10.1186/1471-2105-10-S1-S33
Kamola PJ, Kitson JD, Turner G et al (2015) In silico and in vitro evaluation of exonic and intronic off-target effects form a critical element of therapeutic ASO gapmer optimization. Nucleic Acids Res 43(18):8638–8650. https://doi.org/10.1093/nar/gkv857
Kasuya T, Hori S, Watanabe A et al (2016) Ribonuclease H1-dependent hepatotoxicity caused by locked nucleic acid-modified gapmer antisense oligonucleotides. Sci Rep 6:30377. https://doi.org/10.1038/srep30377
Tolstrup N, Nielsen PS, Kolberg JG et al (2003) OligoDesign: optimal design of LNA (locked nucleic acid) oligonucleotide capture probes for gene expression profiling. Nucleic Acids Res 31(13):3758–3762
Disterer P, Kryczka A, Liu Y et al (2014) Development of therapeutic splice-switching oligonucleotides. Hum Gene Ther 25(7):587–598. https://doi.org/10.1089/hum.2013.234
Kibler-Herzog L, Zon G, Uznanski B et al (1991) Duplex stabilities of phosphorothioate, methylphosphonate, and RNA analogs of two DNA 14-mers. Nucleic Acids Res 19(11):2979–2986
Acknowledgments
This work is supported by the Muscular Dystrophy Canada, the Friends of Garrett Cumming Research Fund, the HM Toupin Neurological Science Research Fund, the Canadian Institutes of Health Research (CIHR), the Alberta Innovates: Health Solutions (AIHS), the Canada Foundation for Innovation (CFI), the Alberta Advanced Education and Technology, and the Women and Children’s Health Research Institute (WCHRI).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Maruyama, R., Yokota, T. (2018). Tips to Design Effective Splice-Switching Antisense Oligonucleotides for Exon Skipping and Exon Inclusion. In: Yokota, T., Maruyama, R. (eds) Exon Skipping and Inclusion Therapies. Methods in Molecular Biology, vol 1828. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-8651-4_5
Download citation
DOI: https://doi.org/10.1007/978-1-4939-8651-4_5
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-8650-7
Online ISBN: 978-1-4939-8651-4
eBook Packages: Springer Protocols