Abstract
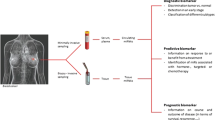
A variety of molecular techniques can be used in order to unravel the molecular composition of cells. In particular, the microarray technology has been used to identify novel biomarkers that may be useful in the diagnosis, prognosis, or treatment of cancer. The microarray technology is ideal for biomarker discovery as it allows for the screening of a large number of molecules at once. In this review, we focus on microRNAs (miRNAs) which are key molecules in cells and regulate gene expression post-transcriptionally. miRNAs are small, single-stranded RNA molecules that bind to complementary mRNAs. Binding of miRNAs to mRNAs leads either to degradation, or translational inhibition of the target mRNA. Roughly one third of all the mRNAs are postulated to be regulated by miRNAs. miRNAs are known to be deregulated in different types of cancer, including breast cancer, and it has been demonstrated that deregulation of several miRNAs can be used as biological markers in cancer. miRNA expression can for example discriminate between normal, benign and malignant breast tissue, and between different breast cancer subtypes.
In the post-genomic era, an important task of molecular biology is to understand gene regulation in the context of biological networks. Because miRNAs have such a pronounced role in cells, it is pivotal to understand the mechanisms that underlie their control, and to identify how miRNAs influence cancer development and progression.
Similar content being viewed by others
References
Crick F (1970) Central dogma of molecular biology. Nature 227(5258):561
Lee RC, Feinbaum RL, Ambros V (1993) The C. elegans heterochronic gene lin-4 encodes small RNAs with antisense complementarity to lin-14. Cell 75(5):843–854
Reinhart BJ, Slack FJ, Basson M, Pasquinelli AE, Bettinger JC, Rougvie AE, Horvitz HR, Ruvkun G (2000) The 21-nucleotide let-7 RNA regulates developmental timing in Caenorhabditis elegans. Nature 403(6772):901–906
Kozomara A, Griffiths-Jones S (2011) miRBase: integrating microRNA annotation and deep-sequencing data. Nucleic Acids Res 39(Database Issue)):D152–D157
Friedman RC, Farh KK-H, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19(1):92–105
Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S et al (2003) The nuclear RNase III Drosha initiates microRNA processing. Nature 425(6956):415–419
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15(8):509–524
Kolb FA, Zhang H, Jaronczyk K, Tahbaz N, Hobman TC, Filipowicz W (2005) Human dicer: purification, properties, and interaction with PAZ PIWI domain proteins. Methods Enzymol 392:316–336
Forman JJ, Legesse-Miller A, Coller HA (2008) A search for conserved sequences in coding regions reveals that the let-7 microRNA targets Dicer within its coding sequence. Proc Natl Acad Sci U S A 105(39):14879–14884
Lytle JR, Yario TA, Steitz JA (2007) Target mRNAs are repressed as efficiently by microRNA-binding sites in the 5′ UTR as in the 3′ UTR. Proc Natl Acad Sci U S A 104(23):9667–9672
Dennis C (2002) The brave new world of RNA. Nature 418(6894):122–124
Sullivan RP, Leong JW, Fehniger TA (2013) MicroRNA regulation of natural killer cells. Front Immunol 4:44
Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JR, Guenther MG, Kumar RM, Murray HL, Jenner RG et al (2005) Core transcriptional regulatory circuitry in human embryonic stem cells. Cell 122(6):947–956
O’Donnell KA, Wentzel EA, Zeller KI, Dang CV, Mendell JT (2005) c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435(7043):839–843
Marson A, Levine SS, Cole MF, Frampton GM, Brambrink T, Johnstone S, Guenther MG, Johnston WK, Wernig M, Newman J et al (2008) Connecting microRNA genes to the core transcriptional regulatory circuitry of embryonic stem cells. Cell 134(3):521–533
Mattick JS, Makunin IV (2006) Non-coding RNA. Hum Mol Genet 15:R17–R29
Czech B, Hannon GJ (2011) Small RNA sorting: matchmaking for Argonautes. Nat Rev Genet 12(1):19–31
Martin G, Schouest K, Kovvuru P, Spillane C (2007) Prediction and validation of microRNA targets in animal genomes. J Biosci 32(6):1049–1052
Thomas M, Lieberman J, Lal A (2010) Desperately seeking microRNA targets. Nat Struct Mol Biol 17(10):1169–1174
Baek D, Villen J, Shin C, Camargo FD, Gygi SP, Bartel DP (2008) The impact of microRNAs on protein output. Nature 455(7209):64–71
Jemal A, Bray F, Center MM, Ferlay J, Ward E, Forman D (2011) Global cancer statistics. CA Cancer J Clin 61(2):69–90
Dupont WD, Page DL (1985) Risk factors for breast cancer in women with proliferative breast disease. N Engl J Med 312(3):146–151
Dupont WD, Page DL, Parl FF, Vnencak-Jones CL, Plummer WD Jr, , Rados MS, Schuyler PA: Long-term risk of breast cancer in women with fibroadenoma. N Engl J Med 1994, 331(1):10–15
McPherson K, Steel CM, Dixon JM (2000) ABC of breast diseases. Breast cancer-epidemiology, risk factors, and genetics. BMJ 321(7261):624–628
Worsham MJ, Raju U, Lu M, Kapke A, Botttrell A, Cheng J, Shah V, Savera A, Wolman SR (2009) Risk factors for breast cancer from benign breast disease in a diverse population. Breast Cancer Res Treat 118(1):1–7
Fitzgibbons PL, Henson DE, Hutter RV (1998) Benign breast changes and the risk for subsequent breast cancer: an update of the 1985 consensus statement. Cancer Committee of the College of American Pathologists. Arch Pathol Lab Med 122(12):1053–1055
McDivitt RW, Stevens JA, Lee NC, Wingo PA, Rubin GL, Gersell D (1992) Histologic types of benign breast disease and the risk for breast cancer. The Cancer and Steroid Hormone Study Group. Cancer 69(6):1408–1414
Cole P, Mark Elwood J, Kaplan SD (1978) Incidence rates and risk factors of benign breast neoplasms. Am J Epidemiol 108(2):112–120
Sgroi DC (2010) Preinvasive breast cancer. Annu Rev Pathol 5:193–221
Johnson K, Sarma D, Hwang ES (2015) Lobular breast cancer series: imaging. Breast Cancer Res 17:94
Perou CM, Sorlie T, Eisen MB, van de Rijn M, Jeffrey SS, Rees CA, Pollack JR, Ross DT, Johnsen H, Akslen LA et al (2000) Molecular portraits of human breast tumours. Nature 406(6797):747–752
Sorlie T, Perou CM, Tibshirani R, Aas T, Geisler S, Johnsen H, Hastie T, Eisen MB, van de Rijn M, Jeffrey SS et al (2001) Gene expression patterns of breast carcinomas distinguish tumor subclasses with clinical implications. Proc Natl Acad Sci U S A 98(19):10869–10874
Gown AM (2008) Current issues in ER and HER2 testing by IHC in breast cancer. Mod Pathol 21(Suppl 2):S8–S15
de Azambuja E, Cardoso F, de Castro G, Colozza M, Mano MS, Durbecq V, Sotiriou C, Larsimont D, Piccart-Gebhart MJ, Paesmans M (2007) Ki-67 as prognostic marker in early breast cancer: a meta-analysis of published studies involving 12 155 patients. Br J Cancer 96(10):1504–1513
van’t Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, Mao M, Peterse HL, van der Kooy K, Marton MJ, Witteveen AT et al (2002) Gene expression profiling predicts clinical outcome of breast cancer. Nature 415(6871):530–536
Enerly E, Steinfeld I, Kleivi K, Aure MR, Leivonen SK, Johnsen H, Kallioniemi O, Kristensen VN, Yakhini Z, Borresen-Dale AL (2010) Molecular characterization of breast cancer subtypes derived from joint analysis of high throughput miRNA and mRNA data. EJC Suppl 8(5):164
Sotiriou C, Neo SY, McShane LM, Korn EL, Long PM, Jazaeri A, Martiat P, Fox SB, Harris AL, Liu ET (2003) Breast cancer classification and prognosis based on gene expression profiles from a population-based study. Proc Natl Acad Sci U S A 100(18):10393–10398
Inic Z, Zegarac M, Inic M, Markovic I, Kozomara Z, Djurisic I, Inic I, Pupic G, Jancic S (2014) Difference between luminal A and luminal B subtypes according to Ki-67, tumor size, and progesterone receptor negativity providing prognostic information. Clin Med Insights Oncol 8:107–111
Subik K, Lee JF, Baxter L, Strzepek T, Costello D, Crowley P, Xing L, Hung MC, Bonfiglio T, Hicks DG et al (2010) The expression patterns of ER, PR, HER2, CK5/6, EGFR, Ki-67 and AR by immunohistochemical analysis in breast cancer cell lines. Breast Cancer (Auckl) 4:35–41
Prat A, Adamo B, Cheang MC, Anders CK, Carey LA, Perou CM (2013) Molecular characterization of basal-like and non-basal-like triple-negative breast cancer. Oncologist 18(2):123–133
Atkinson AJ, Colburn WA, DeGruttola VG, DeMets DL, Downing GJ, Hoth DF, Oates JA, Peck CC, Schooley RT, Spilker BA et al (2001) Biomarkers and surrogate endpoints: preferred definitions and conceptual framework. Clin Pharmacol Therap 69(3):89–95
Sobin LH (2003) TNM: evolution and relation to other prognostic factors. Semin Surg Oncol 21(1):3–7
Karve TM, Cheema AK (2011) Small changes huge impact: the role of protein posttranslational modifications in cellular homeostasis and disease. J Amino Acids 2011:207691
Sharma S, Kelly TK, Jones PA (2010) Epigenetics in cancer. Carcinogenesis 31(1):27–36
Wu W, Zhao S (2013) Metabolic changes in cancer: beyond the Warburg effect. Acta Biochim Biophys Sin Shanghai 45(1):18–26
Calin GA, Dumitru CD, Shimizu M, Bichi R, Zupo S, Noch E, Aldler H, Rattan S, Keating M, Rai K (2002) Frequent deletions and down-regulation of micro-RNA genes miR15 and miR16 at 13q14 in chronic lymphocytic leukemia. Proc Natl Acad Sci U S A 99(24):15524–15529
Calin GA, Sevignani C, Dan Dumitru C, Hyslop T, Noch E, Yendamuri S, Shimizu M, Rattan S, Bullrich F, Negrini M et al (2004) Human microRNA genes are frequently located at fragile sites and genomic regions involved in cancers. Proc Natl Acad Sci U S A 101(9):2999–3004
Iorio MV, Ferracin M, Liu C-G, Veronese A, Spizzo R, Sabbioni S, Magri E, Pedriali M, Fabbri M, Campiglio M et al (2005) MicroRNA gene expression deregulation in human breast cancer. Cancer Res 65(16):7065–7070
Tavazoie SF, Alarcon C, Oskarsson T, Padua D, Wang Q, Bos PD, Gerald WL, Massague J (2008) Endogenous human microRNAs that suppress breast cancer metastasis. Nature 451(7175):147–152
Yan L-X, Huang X-F, Shao Q, Huang MAY, Deng L, Wu Q-L, Zeng Y-X, Shao J-Y (2008) MicroRNA miR-21 overexpression in human breast cancer is associated with advanced clinical stage, lymph node metastasis and patient poor prognosis. RNA 14(11):2348–2360
Castellano L, Giamas G, Jacob J, Coombes RC, Lucchesi W, Thiruchelvam P, Barton G, Jiao LR, Wait R, Waxman J et al (2009) The estrogen receptor-a-induced microRNA signature regulates itself and its transcriptional response. Proc Natl Acad Sci U S A 106(37):15732–15737
Cittelly D, Das P, Spoelstra N, Edgerton S, Richer J, Thor A, Jones F (2010) Downregulation of miR-342 is associated with tamoxifen resistant breast tumors. Mol Cancer 9(1):317
Enerly E, Steinfeld I, Kleivi K, Leivonen S-K, Aure MR, Russnes HG, Rønneberg JA, Johnsen H, Navon R, Rødland E et al (2011) miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One 6(2):e16915
Deng S, Calin GA, Croce CM, Coukos G, Zhang L (2008) Mechanisms of microRNA deregulation in human cancer. Cell Cycle 7(17):2643–2646
Bertoli G, Cava C, Castiglioni I (2015) MicroRNAs: new biomarkers for diagnosis, prognosis, therapy prediction and therapeutic tools for breast cancer. Theranostics 5(10):1122–1143
Nana-Sinkam SP, Croce CM (2013) Clinical applications for microRNAs in cancer. Clin Pharmacol Ther 93(1):98–104
Ouyang M, Li Y, Ye S, Ma J, Lu L, Lv W, Chang G, Li X, Li Q, Wang S et al (2014) MicroRNA profiling implies new markers of chemoresistance of triple-negative breast cancer. PLoS One 9(5):e96228
Dong Y, Wu WK, Wu CW, Sung JJ, Yu J, Ng SS (2011) MicroRNA dysregulation in colorectal cancer: a clinical perspective. Br J Cancer 104(6):893–898
Tumilson CA, Lea RW, Alder JE, Shaw L (2014) Circulating microRNA biomarkers for glioma and predicting response to therapy. Mol Neurobiol 50(2):545–558
Mazan-Mamczarz K, Gartenhaus RB (2013) Role of microRNA deregulation in the pathogenesis of diffuse large B-cell lymphoma (DLBCL). Leuk Res 37(11):1420–1428
Maugeri-Sacca M, Coppola V, Bonci D, De Maria R (2012) MicroRNAs and prostate cancer: from preclinical research to translational oncology. Cancer J 18(3):253–261
Iorio MV, Croce CM (2012) MicroRNA dysregulation in cancer: diagnostics, monitoring and therapeutics. A comprehensive review. EMBO Mol Med 4(3):143–159
Boeri M, Verri C, Conte D, Roz L, Modena P, Facchinetti F, Calabro E, Croce CM, Pastorino U, Sozzi G (2011) MicroRNA signatures in tissues and plasma predict development and prognosis of computed tomography detected lung cancer. Proc Natl Acad Sci U S A 108(9):3713–3718
Cava C, Bertoli G, Ripamonti M, Mauri G, Zoppis I, Della Rosa PA, Gilardi MC, Castiglioni I (2014) Integration of mRNA expression profile, copy number alterations, and microRNA expression levels in breast cancer to improve grade definition. PLoS One 9(5):e97681
Weiler J, Hunziker J, Hall J (2006) Anti-miRNA oligonucleotides (AMOs): ammunition to target miRNAs implicated in human disease? Gene Ther 13(6):496–502
Leivonen SK, Sahlberg KK, Makela R, Due EU, Kallioniemi O, Borresen-Dale AL, Perala M (2014) High-throughput screens identify microRNAs essential for HER2 positive breast cancer cell growth. Mol Oncol 8(1):93–104
Kepp O, Galluzzi L, Lipinski M, Yuan J, Kroemer G (2011) Cell death assays for drug discovery. Nat Rev Drug Discov 10(3):221–237
Riss TL, Moravec RA, Niles AL, Duellman S, Benink HA, Worzella TJ, Minor L (2004) Cell viability assays. In: Sittampalam GS, Coussens NP, Brimacombe K, Grossman A, Arkin M, Auld D, Austin C, Bejcek B, Glicksman M, Inglese J et al (eds) Assay guidance manual. Eli Lilly & Company, Bethesda, MD
Zhu Q, Wong AK, Krishnan A, Aure MR, Tadych A, Zhang R, Corney DC, Greene CS, Bongo LA, Kristensen VN et al (2015) Targeted exploration and analysis of large cross-platform human transcriptomic compendia. Nat Methods 12(3):211–214
Enright A, John B, Gaul U, Tuschl T, Sander C, Marks D (2003) MicroRNA targets in Drosophila. Genome Biol 5(1):R1
John B, Enright AJ, Aravin A, Tuschl T, Sander C, Marks DS (2004) Human microRNA targets. PLoS Biol 2(11):e363
Betel D, Wilson M, Gabow A, Marks DS, Sander C (2008) The microRNA.org resource: targets and expression. Nucleic Acids Res 36(Database Issue):D149–D153
Lewis BP, Burge CB, Bartel DP (2005) Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell 120(1):15–20
Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB (2003) Prediction of mammalian microRNA targets. Cell 115(7):787–798
Krek A, Grun D, Poy MN, Wolf R, Rosenberg L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M et al (2005) Combinatorial microRNA target predictions. Nat Genet 37(5):495–500
Grun D, Wang YL, Langenberger D, Gunsalus KC, Rajewsky N (2005) microRNA target predictions across seven Drosophila species and comparison to mammalian targets. PLoS Comput Biol 1(1):e13
Lall S, Grun D, Krek A, Chen K, Wang YL, Dewey CN, Sood P, Colombo T, Bray N, Macmenamin P et al (2006) A genome-wide map of conserved microRNA targets in C. elegans. Curr Biol 16(5):460–471
Maragkakis M, Alexiou P, Papadopoulos GL, Reczko M, Dalamagas T, Giannopoulos G, Goumas G, Koukis E, Kourtis K, Simossis VA et al (2009) Accurate microRNA target prediction correlates with protein repression levels. BMC Bioinformatics 10:295
Paraskevopoulou MD, Georgakilas G, Kostoulas N, Vlachos IS, Vergoulis T, Reczko M, Filippidis C, Dalamagas T, Hatzigeorgiou AG (2013) DIANA-microT web server v5.0: service integration into miRNA functional analysis workflows. Nucleic Acids Res 41(W1):W169–W173
Ekimler S, Sahin K (2014) Computational methods for microRNA target prediction. Genes (Basel) 5(3):671–683
Hanahan D, Weinberg RA (2011) Hallmarks of cancer: the next generation. Cell 144(5):646–674
Hyman E, Kauraniemi P, Hautaniemi S, Wolf M, Mousses S, Rozenblum E, Ringnér M, Sauter G, Monni O, Elkahloun A et al (2002) Impact of DNA amplification on gene expression patterns in breast cancer. Cancer Res 62(21):6240–6245
Bergamaschi A, Kim YH, Wang P, Sørlie T, Hernandez-Boussard T, Lonning PE, Tibshirani R, Børresen-Dale A-L, Pollack JR (2006) Distinct patterns of DNA copy number alteration are associated with different clinicopathological features and gene-expression subtypes of breast cancer. Genes Chromosom Cancer 45(11):1033–1040
Aure MR, Leivonen SK, Fleischer T, Zhu Q, Overgaard J, Alsner J, Tramm T, Louhimo R, Alnæs GI, Perälä M, Busato F, Touleimat N, Tost J, Børresen-Dale AL, Hautaniemi S, Troyanskaya OG, Lingjærde OC, Sahlberg KK, Kristensen VN (2013) Individual and combined effects of DNA methylation and copy number alterations on miRNA expression in breast tumors. Genome Biol 14(11):R126
Lahti L, Schäfer M, Klein H-U, Bicciato S, Dugas M (2012) Cancer gene prioritization by integrative analysis of mRNA expression and DNA copy number data: a comparative review. Brief Bioinform 14(1):27–35
Louhimo R, Lepikhova T, Monni O, Hautaniemi S (2012) Comparative analysis of algorithms for integration of copy number and expression data. Nat Methods 9(4):351–355
Huang N, Shah PK, Li C (2011) Lessons from a decade of integrating cancer copy number alterations with gene expression profiles. Brief Bioinform 13(3):305–316
Zhang S, Liu C-C, Li W, Shen H, Laird PW, Zhou XJ (2012) Discovery of multi-dimensional modules by integrative analysis of cancer genomic data. Nucleic Acids Res 40(19):9379–9391
McDermott JE, Costa M, Janszen D, Singhal M, Tilton SC (2010) Separating the drivers from the driven: integrative network and pathway approaches aid identification of disease biomarkers from high-throughput data. Dis Markers 28(4):253–266
Baudot A, Real FX, Izarzugaza JMG, Valencia A (2009) From cancer genomes to cancer models: bridging the gaps. EMBO Rep 10(4):359–366
Chin L, Hahn WC, Getz G, Meyerson M (2011) Making sense of cancer genomic data. Genes Dev 25(6):534–555
Dvinge H, Git A, Graf S, Salmon-Divon M, Curtis C, Sottoriva A, Zhao Y, Hirst M, Armisen J, Miska EA et al (2013) The shaping and functional consequences of the microRNA landscape in breast cancer. Nature 497(7449):378–382
Curtis C, Shah SP, Chin SF, Turashvili G, Rueda OM, Dunning MJ, Speed D, Lynch AG, Samarajiwa S, Yuan Y et al (2012) The genomic and transcriptomic architecture of 2,000 breast tumours reveals novel subgroups. Nature 486(7403):346–352
Hernández Patiño CE, Jaime-Muñoz G, Resendis-Antonio O (2013) Systems biology of cancer: moving toward the integrative study of the metabolic alterations in cancer cells. Front Physiol 3:481
The Cancer Genome Atlas Network (2012) Comprehensive molecular portraits of human breast tumours. Nature 490(7418):61–70
Muniategui A, Pey J, Planes FJ, Rubio A (2012) Joint analysis of miRNA and mRNA expression data. Brief Bioinform 14(3):263–278
Chi SW, Zang JB, Mele A, Darnell RB (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460(7254):479–486
Aure MR, Jernstrom S, Krohn M, Vollan H, Due E, Rodland E, Karesen R, Ram P, Lu Y, Mills G et al (2015) Integrated analysis reveals microRNA networks coordinately expressed with key proteins in breast cancer. Genome Med 7(1):21
Hertel J, Lindemeyer M, Missal K, Fried C, Tanzer A, Flamm C, Hofacker I, Stadler P, Students of Bioinformatics Computer Labs 2004 and 2005 (2006) The expansion of the metazoan microRNA repertoire. BMC Genomics 7:25
Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ (2008) miRBase: tools for microRNA genomics. Nucleic Acids Res 36(Database issue):D154–D158
Inui M, Martello G, Piccolo S (2010) MicroRNA control of signal transduction. Nat Rev Mol Cell Biol 11(4):252–263
Bentwich I (2005) Prediction and validation of microRNAs and their targets. FEBS Lett 579(26):5904–5910
Patnaik SK, Dahlgaard J, Mazin W, Kannisto E, Jensen T, Knudsen S, Yendamuri S (2012) Expression of microRNAs in the NCI-60 cancer cell-lines. PLoS One 7(11):e49918
Lu J, Getz G, Miska EA, Alvarez-Saavedra E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA et al (2005) MicroRNA expression profiles classify human cancers. Nature 435(7043):834–838
Selbach M, Schwanhausser B, Thierfelder N, Fang Z, Khanin R, Rajewsky N (2008) Widespread changes in protein synthesis induced by microRNAs. Nature 455(7209):58–63
Creixell P, Schoof EM, Erler JT, Linding R (2012) Navigating cancer network attractors for tumor-specific therapy. Nat Biotechnol 30(9):842–848
Avraham R, Yarden Y (2012) Regulation of signalling by microRNAs. Biochem Soc Trans 40(1):26–30
Mitchell PS, Parkin RK, Kroh EM, Fritz BR, Wyman SK, Pogosova-Agadjanyan EL, Peterson A, Noteboom J, O'Briant KC, Allen A et al (2008) Circulating microRNAs as stable blood-based markers for cancer detection. Proc Natl Acad Sci U S A 105(30):10513–10518
Haakensen VD, Nygaard V, Greger L, Aure MR, Fromm B, Bukholm IR, Luders T, Chin SF, Git A, Caldas C et al (2016) Subtype-specific micro-RNA expression signatures in breast cancer progression. Int J Cancer 139(5):1117–1128
Lesurf R, Aure MR, Mork HH, Vitelli V, Oslo Breast Cancer Research Consortium, Lundgren S, Borresen-Dale AL, Kristensen V, Warnberg F, Hallett M et al (2016) Molecular features of subtype-specific progression from ductal carcinoma in situ to invasive breast cancer. Cell Rep 16(4):1166–1179
Tahiri A, Leivonen SK, Luders T, Steinfeld I, Aure MR, Geisler J, Makela R, Nord S, Riis MLH, Yakhini Z et al (2014) Deregulation of cancer-related miRNAs is a common event in both benign and malignant human breast tumors. Carcinogenesis 35(1):76–85
Callari M, Dugo M, Musella V, Marchesi E, Chiorino G, Grand MM, Pierotti MA, Daidone MG, Canevari S, De Cecco L (2012) Comparison of microarray platforms for measuring differential microRNA expression in paired normal/cancer colon tissues. PLoS One 7(9):e45105
Backes C, Sedaghat-Hamedani F, Frese K, Hart M, Ludwig N, Meder B, Meese E, Keller A (2016) Bias in high-throughput analysis of miRNAs and implications for biomarker studies. Anal Chem 88(4):2088–2095
Meiri E, Mueller WC, Rosenwald S, Zepeniuk M, Klinke E, Edmonston TB, Werner M, Lass U, Barshack I, Feinmesser M et al (2012) A second-generation microRNA-based assay for diagnosing tumor tissue origin. Oncologist 17(6):801–812
Garzon R, Marcucci G, Croce CM (2010) Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov 9(10):775–789
Thorsen SB, Obad S, Jensen NF, Stenvang J, Kauppinen S (2012) The therapeutic potential of microRNAs in cancer. Cancer J 18(3):275–284
Aagaard L, Rossi JJ (2007) RNAi therapeutics: principles, prospects and challenges. Adv Drug Deliv Rev 59(2–3):75–86
Yan LX, Wu QN, Zhang Y, Li YY, Liao DZ, Hou JH, Fu J, Zeng MS, Yun JP, Wu QL et al (2011) Knockdown of miR-21 in human breast cancer cell lines inhibits proliferation, in vitro migration and in vivo tumor growth. Breast Cancer Res 13(1):R2
Lujambio A, Lowe SW (2012) The microcosmos of cancer. Nature 482(7385):347–355
Acknowledgments
Parts of this review have been part of two doctoral theses from the University of Oslo, Norway, under the supervision of V.N.K.: one of M.R.A., fellow of the Research Council of Norway, and one of A.T., fellow of the South-Eastern Norway Regional Health Authority. Both are at present postdoctoral fellows of the South-Eastern Norway Regional Health Authority.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Tahiri, A., Aure, M.R., Kristensen, V.N. (2018). MicroRNA Networks in Breast Cancer Cells. In: von Stechow, L. (eds) Cancer Systems Biology. Methods in Molecular Biology, vol 1711. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7493-1_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7493-1_4
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7492-4
Online ISBN: 978-1-4939-7493-1
eBook Packages: Springer Protocols