Abstract
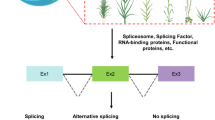
Alternative splicing (AS) promotes transcriptome and proteome diversity in plants, which influences growth and development, and host responses to stress. Advancements in next-generation sequencing, bioinformatics, and computational biology tools have allowed biologists to investigate AS landscapes on a genome-wide scale in several plant species. Furthermore, the development of Brachypodium distachyon (Brachypodium) as a model system for grasses has facilitated comparative studies of AS within the Poaceae. These analyses revealed a plethora of genes in several biological processes that are alternatively spliced and identified conserved AS patterns among monocot and dicot plants. In this chapter, using a Brachypodium-virus pathosystem as a research template, we provide an overview of genomic and bioinformatic tools that can be used to investigate constitutive and alternative splicing in plants.
Similar content being viewed by others
References
Staiger D, Brown JWS (2013) Alternative splicing at the intersection of biological timing, development, and stress responses. Plant Cell 25(10):3640–3656. doi:10.1105/tpc.113.113803
Reddy ASN, Marquez Y, Kalyna M, Barta A (2013) Complexity of the alternative splicing landscape in plants. Plant Cell 25(10):3657–3683. doi:10.1105/tpc.113.117523
Dillies M-A, Rau A, Aubert J, Hennequet-Antier C, Jeanmougin M, Servant N, Keime C, Marot G, Castel D, Estelle J, Guernec G, Jagla B, Jouneau L, Laloë D, Le Gall C, Schaëffer B, Le Crom S, Guedj M, Jaffrézic F (2013) A comprehensive evaluation of normalization methods for Illumina high-throughput RNA sequencing data analysis. Brief Bioinform 14(6):671–683. doi:10.1093/bib/bbs046
Katz Y, Wang ET, Airoldi EM, Burge CB (2010) Analysis and design of RNA sequencing experiments for identifying isoform regulation. Nat Methods 7(12):1009–1015. doi:10.1038/nmeth.1528
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25(9):1105–1111. doi:10.1093/bioinformatics/btp120
Mandadi KK, Scholthof K-BG (2015) Genomic architecture and functional relationships of intronless, constitutively- and alternatively-spliced genes in Brachypodium distachyon. Plant Signal Behav 10(8):e1042640. doi:10.1080/15592324.2015.1042640
Mandadi KK, Scholthof K-BG (2015) Genome-wide analysis of alternative splicing landscapes modulated during plant-virus interactions in Brachypodium distachyon. Plant Cell 27:71–85. doi:10.1105/tpc.114.133991
Mandadi KK, Pyle JD, Scholthof K-BG (2015) Characterization of SCL33 splicing patterns during diverse virus infections in Brachypodium distachyon. Plant Signal Behav 10(8):e1042641. doi:10.1080/15592324.2015.1042641
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7(3):562–578. doi:10.1038/nprot.2012.016
Liu R, Loraine AE, Dickerson JA (2014) Comparisons of computational methods for differential alternative splicing detection using RNA-seq in plant systems. BMC Bioinformatics 15(1):364
Thomas J, Palusa SG, Prasad KVSK, Ali GS, Surabhi G-K, Ben-Hur A, Abdel-Ghany SE, Reddy ASN (2012) Identification of an intronic splicing regulatory element involved in auto-regulation of alternative splicing of SCL33 pre-mRNA. Plant J 72(6):935–946. doi:10.1111/tpj.12004
Goff SA, Vaughn M, McKay S, Lyons E, Stapleton AE, Gessler D, Matasci N, Wang L, Hanlon M, Lenards A, Muir A, Merchant N, Lowry S, Mock S, Helmke M, Kubach A, Narro M, Hopkins N, Micklos D, Hilgert U, Gonzales M, Jordan C, Skidmore E, Dooley R, Cazes J, McLay R, Lu Z, Pasternak S, Koesterke L, Piel WH, Grene R, Noutsos C, Gendler K, Feng X, Tang C, Lent M, Kim S-J, Kvilekval K, Manjunath BS, Tannen V, Stamatakis A, Sanderson M, Welch SM, Cranston K, Soltis P, Soltis D, O’Meara B, Ane C, Brutnell T, Kleibenstein DJ, White JW, Leebens-Mack J, Donoghue MJ, Spalding EP, Vision TJ, Myers CR, Lowenthal D, Enquist BJ, Boyle B, Akoglu A, Andrews G, Ram S, Ware D, Stein L, Stanzione D (2011) The iPlant Collaborative: cyberinfrastructure for plant biology. Front Plant Sci 2(34). doi:10.3389/fpls.2011.00034
Goecks J, Nekrutenko A, Taylor J, Team TG (2010) Galaxy: a comprehensive approach for supporting accessible, reproducible, and transparent computational research in the life sciences. Genome Biol 11(8):R86. doi:10.1186/gb-2010-11-8-r86
Loraine AE, McCormick S, Estrada A, Patel K, Qin P (2013) RNA-seq of Arabidopsis pollen uncovers novel transcription and alternative splicing. Plant Physiol 162(2):1092–1109. doi:10.1104/pp.112.211441
Biswas S, Agrawal YN, Mucyn TS, Dangl JL, Jones CD (2013) Biological averaging in RNA-Seq. arXiv 1309.0670
Sims D, Sudbery I, Ilott NE, Heger A, Ponting CP (2014) Sequencing depth and coverage: key considerations in genomic analyses. Nat Rev Genet 15(2):121–132. doi:10.1038/nrg3642
Patel RK, Jain M (2012) NGS QC Toolkit: a toolkit for quality control of next generation sequencing data. PLoS One 7(2):e30619
Langmead B, Trapnell C, Pop M, Salzberg SL (2009) Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol 10(3):R25
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL (2013) TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol 14(4):R36
Trapnell C, Hendrickson DG, Sauvageau M, Goff L, Rinn JL, Pachter L (2013) Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol 31(1):46–53. doi:10.1038/nbt.2450
Pertea M, Pertea GM, Antonescu CM, Chang T-C, Mendell JT, Salzberg SL (2015) StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33(3):290–295
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28(5):511–515. doi:10.1038/nbt.1621
Thorvaldsdóttir H, Robinson JT, Mesirov JP (2013) Integrative Genomics Viewer (IGV): high-performance genomics data visualization and exploration. Brief Bioinform 14(2):178–192. doi:10.1093/bib/bbs017
Foissac S, Sammeth M (2015) Analysis of alternative splicing events in custom gene datasets by AStalavista. Methods Mol Biol 1269:379–392. doi:10.1007/978-1-4939-2291-8_24
Foissac S, Sammeth M (2007) ASTALAVISTA: dynamic and flexible analysis of alternative splicing events in custom gene datasets. Nucleic Acids Res 35(suppl 2):W297–W299. doi:10.1093/nar/gkm311
Brooks AN, Yang L, Duff MO, Hansen KD, Park JW, Dudoit S, Brenner SE, Graveley BR (2011) Conservation of an RNA regulatory map between Drosophila and mammals. Genome Res 21(2):193–202
Katz Y, Wang ET, Stilterra J, Schwartz S, Wong B, Thorvaldsdóttir H, Robinson JT, Mesirov JP, Airoldi EM, Burge CB (2014) Sashimi plots: quantitative visualization of alternative isoform expression from RNA-seq data. arXiv:1306.3466. doi:10.1101/002576
Acknowledgment
This study was supported by funds from USDA-NIFA-AFRI (2016-67013-24738) to K-B.G.S. and K.K.M., Texas A&M AgriLife Research Bioenergy/Bioproducts Grant (124738-96210) to K.K.M.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2018 Springer Science+Business Media LLC
About this protocol
Cite this protocol
Irigoyen, S., Bedre, R.H., Scholthof, KB.G., Mandadi, K.K. (2018). Genomic Approaches to Analyze Alternative Splicing, A Key Regulator of Transcriptome and Proteome Diversity in Brachypodium distachyon . In: Sablok, G., Budak, H., Ralph, P. (eds) Brachypodium Genomics. Methods in Molecular Biology, vol 1667. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-7278-4_7
Download citation
DOI: https://doi.org/10.1007/978-1-4939-7278-4_7
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-7276-0
Online ISBN: 978-1-4939-7278-4
eBook Packages: Springer Protocols