Abstract
MicroRNAs are short noncoding, ~22-nucleotide RNAs that regulate protein abundance. The growth in our understanding of this class of RNAs has been rapid since their discovery just over a decade ago. We now appreciate that miRNAs are deeply embedded within the genetic networks that control basic features of metazoan cells including their identity, metabolism, and reproduction. The Drosophila melanogaster model system has made and will continue to make important contributions to this analysis. Intended as an introductory overview, here we review the current methods and resources available for functional analysis of fly miRNAs for those interested in performing this type of analysis.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
This chapter reviews molecular and genetic methods and resources for the analysis of microRNA (miRNA) function in the Drosophila melanogaster (D. melanogaster) model system. Biochemical analysis of the fly system has also provided critical insights into the upstream pathways that control miRNA production. We start with a brief summary of fly miRNA biogenesis, but see two excellent recent reviews [1, 2] and their references for more detailed descriptions of this aspect of fly miRNAs.
Mature miRNAs are ~22 nucleotides (nts) long but are derived from slightly longer, ~70-nt RNAs that fold into hairpin structures known as pre-miRNAs. These pre-miRNAs are transcribed within even longer primary transcripts, or pri-miRNAs, that can be hundreds or thousands of base-pairs long. Pri-miRNAs are cleaved by a ribonuclease complex containing two proteins , Drosha and Pahsa, to release pre-miRNAs, which are exported from the nucleus and subsequently cut by a second ribonuclease Dicer. This cleavage releases two different short RNAs coming from either the 5′ or 3′ arm of the hairpin. Current naming conventions distinguish these related miRNAs with -5p or -3p suffixes, respectively, [3] and usually one of these predominates while the other is degraded. Once released, the active miRNA incorporates into a protein complex that includes Argonaute1 (Ago1) and directs this silencing complex to partially complementary sequences of target mRNAs, usually to quench their translation or promote their degradation. Below, we summarize methods to identify miRNAs as well as characterize their expression patterns, targets, and in vivo functions.
2 Identification
The previous version of this chapter described early efforts to identify 78 D. melanogaster miRNAs via molecular cloning and sequencing of small RNAs as well as in silico predictions [4]. Since that chapter was published in 2008, the construction and deeper sequencing of numerous small RNA libraries as well as the detailed characterization of 12 genomes of related Drosophila species [5] has identified many additional miRNAs. The current tally of fly miRNA hairpins is 256 [3] and these include both canonical miRNAs as well as mirtrons, which are derived from spliced introns and follow a slightly altered biogenesis pathway [5–11]. The rate at which new miRNAs are discovered has decreased in recent years, suggesting that the total number of fly miRNAs has reached its upper limit [11]. This section summarizes recent miRNA identification efforts as well as some resulting insights into miRNA biogenesis and modifications. Please see ref. [1] and [2] for more detailed discussion of miRNA biogenesis in flies and other models.
Groups associated with the Model Organism Encyclopedia of DNA Elements (modENCODE) project have assembled a comprehensive catalog of the small RNA contingent of the fly transcriptome. Collectively, the analysis of roughly 1.5 billion sequencing reads identified 146 new miRNAs [6, 7, 10–13]. These reads were obtained from RNA libraries generated from various developmental stages, tissues, mutants, cell lines , and protein immunoprecipitates (IP). For example, IP of Ago1 from total tissue extract enriches the isolation of miRNAs engaged in silencing since Ago1 is a critical component of the miRNA effector complex. Identified miRNAs originated from both sense and antisense transcription units, intergenic regions, introns, intron–exon junctions as well as occasionally from the untranslated and coding regions of genes [7]. In addition to cataloguing processed small RNAs, Graveley et al. also annotated a set of 23 primary miRNA transcripts using deep sequencing and cap analysis of gene expression (CAGE). The magnitude of these deep sequencing efforts has provided a detailed picture of the small RNA landscape, capturing not only abundantly produced mature miRNAs but also byproducts generated during miRNA processing. As outlined below, analysis of these byproducts has revealed unanticipated aspects of miRNA production.
2.1 Mirtrons
Mirtrons were identified as hairpins whose 5′ and 3′ ends coincide with intron termini [9]. These hairpins are released by splicing, bypassing the usual miRNA processing by Drosha and Pasha. Initial analysis of approximately one million reads identified 14 mirtrons and subsequent analysis of 17 additional million reads identified 5 more [6, 9]. A computational machine learning approach using the original 14 mirtrons as a training set yielded 51 additional mirtron candidates, and six of these were found in modENCODE small RNA data sets [8]. The total number of fly mirtrons is currently estimated to be ~30 [8].
2.2 IsomiRs and miRNA Modifications
Mature miRNAs can have multiple isoforms, termed isomiRs, due to heterogeneities that arise at both 5′ and 3′ termini [7, 10, 14]. Sequence heterogeneity at the 5′ end is likely due to alternative processing by Drosha and/or Dicer [7]. A prominent example is miR-210: two versions of miR-210—one with one additional 5′ nucleotide—are equally represented in RNA libraries [10]. Modifications at the 3′ end are due to untemplated additions, including uridylation and adenylation, as well as trimming [1, 2]. The exonuclease Nibbler, for example, is responsible for 3′ trimming of miR-34 mature sequence, resulting in multiple miR-34 isoforms [14, 15]. The consequences of these modifications are not known, but likely impact either miRNA stability and/or miRNA–target interactions.
In addition, miRNA sequences can be edited post-transcriptionally by the Adenosine Deaminase acting on RNA (ADAR) enzyme. Editing events can change miRNA seed sequences, affect processing by Drosha/Pasha/Dicer, and even alter Ago-sorting preference [7]. Deep sequencing analysis of processed miRNAs identified three mature miRNAs, miR-100, miR-971, and miR-33*, that frequently contained A to G nucleotide changes. Subsequent analysis of miR-100 and its co-transcribed neighbors, let-7 and miR-125, identified additional editing events in the hairpins of all three miRNAs and found that some of these editing events control the differential expression of the three mature miRNAs in vitro and in vivo [16].
2.3 Additional miRNA Hairpin Products
Additional products of miRNA hairpins are functional including loop sequences. For example, miR-34 and miR-317 hairpin loop sequences were recovered in IPs of the miRNA effector Ago1 [17]. These loop sequences were generated during miRNA biogenesis and, consistent with their association with Ago1, were functional and repressed the expression of reporter transgenes containing complementary sequences. The in vivo function of these hairpin RNAs as well as the byproducts of other miRNA hairpins remains to be determined.
3 Spatiotemporal Detection
Knowledge of the expression of miRNAs is crucial for understanding the functional roles of miRNAs. This section outlines methods that have been utilized for detecting the spatiotemporal expression patterns of fly miRNAs.
3.1 Transcriptional Reporters
The expression pattern of primary miRNA transcripts can be determined by generating transgenic lines containing promoter/enhancer fragments fused to reporters such as GFP or LacZ. Such an approach has been utilized to determine in vivo expression of miR-1 [18–20], miR-124 [21], miR-278 [22], miR-309/3/286/4/5/6-1/6-2/6-3 [18], and the let-7-Complex (let-7-C) locus [23]. Designing transcriptional reporters requires prior knowledge of the transcription start site that can be mapped using 5′RACE. This helps preclude open reading frames present in the 5′UTR of primary miRNA transcripts in the reporter and its subsequent degradation by nonsense-mediated decay.
3.2 Sensors
“MiRNA sensors” are ubiquitously expressed transgene reporters (GFP or luciferase) that contain one or more perfectly complementary miRNA binding sites in the 3′UTR. The miRNA sensors function like a target for miRNA-mediated destruction via the RNA interference machinery, and hence their expression is reduced in regions of high miRNA activity. This technique has been successfully used to report functional read-outs of miRNAs as well as in genetic screens for functional characterization of the miRNA biogenesis pathway [16, 24–31]. One limitation of this technique is reporter protein perdurance, which could mask the activity of dynamically expressed miRNAs. More recently, cleavable sensor oligonucleotides have been described that report miRNA expression in live mammalian cells [32]. Use of these activatable sensors will allow researchers to report dynamics of miRNA-mediated regulation in diverse contexts such as cell fate determination.
3.3 In Situ Hybridization
In situ hybridization (ISH) is an invaluable method to directly visualize mRNAs at a cellular resolution in whole organisms and tissues as well as tissue sections. This hybridization-based technique has been utilized to study the expression pattern of both primary as well as processed miRNAs in specific cell types. The most popular ISH techniques involve the use of alkaline phosphatase or fluorophore-conjugated probes and fluorescent tyramide signal amplification (TSA)-based procedures [33]. Primary miRNA transcripts have been detected with antisense probes corresponding to the genomic DNA surrounding the miRNA hairpin precursor. This approach was successfully used to monitor the dynamic expression of pri-miRNAs involved in patterning of D. melanogaster embryos [34–36]. Since processing of a primary miRNA transcript is a regulated event, the temporal and spatial expression of the parental primary transcripts may or may not coincide with the expression of the miRNA that it generates [37]. Thus, several advances have been made to facilitate ISH detection of mature miRNAs. Labeled locked nucleic acid (LNA) probes and either a colorimetric or fluorescent method for probe detection have been successfully used to detect miRNA expression in D. melanogaster , including let-7, miR-1, miR-7, miR-10, miR-34, miR-252, miR-956, and miR-980 [19, 38–43]. LNA probes are a class of RNA analogs in which the 2′oxygen and the 4′carbon positions in the ribose ring are connected or “locked” to create increased stability relative to DNA or RNA when they are base-paired with complementary DNA or RNA. The development of LNA probes has circumvented the limitations associated with detection of mature miRNAs that have a lower hybridization potential owing to their short length. More recently, immunofluorescence coupled to in situ hybridization (IF-ISH) protocols for dual detection of protein and miRNA have been described for D. melanogaster tissues [33]. These will likely allow simultaneous detection of miRNAs in specific cell types (for e.g. stem cells ) that can be marked and visualized with antibodies in specific tissues.
3.4 Northern Blot Analysis
Northern blot analysis has been used extensively for the detection of mature and precursor miRNAs in diverse tissues and cell lines [22, 23]. The technique involves running total RNA on a denaturing polyacrylamide gel followed by its transfer to a nitrocellulose membrane. The RNA is fixed onto the membrane by UV crosslinking and incubated with antisense radiolabeled or fluorescent probes. Though Northern analysis does not require any special equipment and also provides high-quality confirmation, the low level of sensitivity, large amounts of RNA required as well as the time-consuming protocols have resulted in improvisation of the traditional protocol [44]. For instance, the use of LNA-modified northern probes that exhibit higher thermal stability and show improved hybridization properties has increased the sensitivity of the Northern blotting technique and allowed more specific detection of miRNAs [45, 46].
3.5 Quantitative Real-Time PCR
Quantitative real-time PCR (qRT-PCR) is the most widely used method to detect and quantify miRNA levels. This technique relies on reverse transcription of the miRNA followed by qPCR with real-time monitoring. Some of the limitations of miRNA reverse transcription are (1) the short length of the miRNA, (2) the presence of the mature miRNA sequence in the precursor and primary miRNA transcript, and (3) the lack of common sequence features like poly (A) tails.
Two main methods have been developed to reverse transcribe miRNAs: universal and miRNA-specific reverse transcription . In the first approach, the 3′end of all miRNAs are elongated with a poly(A) tail using an E. coli poly(A) polymerase (miRCURY, Exiqon). This is followed by reverse transcription with an oligo(dT) primer containing a universal primer-binding sequence. PCR amplification is enabled by a miRNA-specific forward primer and a reverse primer that anneals to the 3′portion of the miRNA as well as the poly(A) tail with SYBR green dye as the detector. The second approach utilizes a stem-loop primer for reverse transcription (Taqman, Life Technologies) (Fig. 1a, d) and has been useful for detection of D. melanogaster miRNAs [16, 47, 48]. The stem-loop sequence in the primer reduces annealing to the primary and precursor miRNA, thereby increasing specificity of the assay. This assay method is not affected by genomic DNA contamination and is able to discriminate between miRNAs that differ by just one nucleotide.
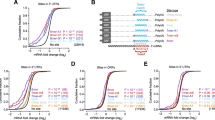
Methods for profiling miRNAs. (a) TaqMan quantitative real-time PCR (qRT-PCR). The miRNA in the RNA sample is reverse transcribed with a stem-loop primer that base-pairs to the 3′end of the miRNA. The resulting cDNA is amplified in the presence of a miRNA-specific forward and reverse primer in presence of a miRNA-specific Taqman probe. The Taqman probe has a fluorescent reporter linked to its 5′ end and a nonfluorescent quencher at the 3′ end of the probe. During PCR, the Taqman probe anneals specifically to the sequence between forward and primer sites. As the DNA polymerase proceeds along the template, it cleaves the Taqman probe that is hybridized to the template. This cleavage separates the fluorescent reporter from the quencher dye, resulting in an increase in fluorescence of the reporter. Taqman miRNA assays are available in an array format and can be used for high-throughput analysis of miRNAs. (b) MiRNA microarrays. miRNAs in an RNA sample are fluorescently labeled and hybridized to DNA-based capture probes (with LNA-modified bases) spotted on arrays. The microarrays are washed and scanned to detect fluorescence intensity. (c) RNA-seq. A cDNA library is generated by reverse transcription of miRNAs in the sample RNA. The cDNAs in the library are ligated to adapter molecules that allow the library to be affixed to a solid phase. The Illumina/Solexa technology utilizes a “bridged amplification” that occurs on the surface of the flow cell. After amplification, the flow cell is exposed to reagents required for sequencing. (d) Table comparing different available methods for miRNA profiling
Another advantage of this approach is that it can be easily scaled up for high-throughput analysis. For example, commercially available customizable plates and microfluidic cards can be designed to assay entire sets of miRNAs (Fig. 1a, d). Some of the advantages of this assay system include high sensitivity and a wide dynamic range. qRT-PCR is the only method that can provide absolute miRNA quantitation and is accomplished by generating standard curves from synthetic oligonucleotides of known concentration [49].
3.6 miRNA Microarrays
Microarray technology is a high-throughput approach to monitor expression of several miRNAs in a single experiment. This technique involves purification of small RNAs from cells and tissues followed by fluorescent labeling of the enriched small RNA fraction (Fig. 1b, d). The labeled RNA is hybridized to arrays that are spotted with the appropriate high-affinity probes specific to mature RNAs. Microarray scanners are used to detect the spot intensity of the double-stranded fragments. Either constitutively expressed miRNAs or U6 snRNA or tRNA is used for data normalization. Although microarray analysis may require optimization to determine the best hybridization condition, it allows the simultaneous comparison of expression levels of several miRNAs in a single experiment at a low cost. The expression profiles obtained from the microarray analysis need further validation by other quantitative techniques like Northern analysis or real-time PCR. This approach has been utilized to determine age-modulated expression of miRNAs in adult fly brains [50], whole animals [51], and during embryogenesis in flies overexpressing dMyc [52].
3.7 Deep Sequencing
In the past couple years, next-generation sequencing technology (NGS) has allowed high-throughput detection of small RNAs with a high degree of reliability [53]. NGS technology allows sequencing of large numbers of DNA fragments in parallel, producing millions of short reads in a single run of an automated sequencer [54, 55]. The most widely adopted NGS platform that has been successfully used in the D. melanogaster model is the Illumina/Solexa technology [7, 11, 56] (Fig. 1c, d). One of the advantages of using RNA sequencing (RNA-seq) over microarray profiling is that it allows discovery of novel miRNAs. Other advantages include the extremely high level of sensitivity and resolution that allows detection of miRNAs that are very similar to one another, including isomiRs that differ by a single base. However, the analysis of data obtained from NGS platforms requires substantial computational support and it cannot provide an absolute quantitative view of these transcripts. For a given sample analyzed by RNA-seq, miRNA quantitation is expressed as a number relative to the total number of sequence reads for the sample, thus comparisons between samples with high variance in miRNA expression are not reliable owing to the difference in the number of reads. Furthermore, the number of reads obtained for any miRNA may not necessarily correlate with its actual abundance owing to the biases introduced during sample preparation and sequencing.
4 Targets
MiRNAs carry out their biological functions by regulating the expression of target mRNAs. Estimates suggest that an individual miRNA can regulate hundreds of targets and the fly has been a premier system for ascertaining which of its targets is biologically relevant. This section summarizes methods for identifying and verifying miRNA targets.
4.1 Bioinformatic Strategies
A first step in identifying miRNA targets is the use of target prediction algorithms. The most common of these include TargetScan, Pictar, and miRanda [57]. These programs search curated sets of 3′UTR sequences for likely miRNA-binding sites based on various criteria including sequence complementarity, duplex free energy, and conservation [43, 57–62]. A frequently imposed guideline requires the presence of six contiguous base-pairs in the target that are complementary to the 5′ end, or “seed” region, of the miRNA [57]. Recently, Chi et al. proposed an additional guideline for target recognition that permits the presence of a “nucleation bulge” in the seed region [63, 64]. This interaction requires only five consecutive base-pairings at the position 2–6 of the miRNA to form a sufficiently stable duplex. Incorporation of this guideline into future algorithms will likely improve predictions.
One current limitation of these commonly used algorithms is that they are not updated frequently with new 3′UTRs. They therefore rely on outdated 3′UTR information and are missing more recently identified alternative 3′UTR, isoform-specific 3′UTR, or extended 3′UTR sequences [65]. Direct integration of revised annotations into the target prediction tools should address this problem. In addition, manually curated 3′UTR sequences can be searched for predicted miRNA binding sites using the RNAhybrid program [66].
In addition, the high rates of false discovery of target prediction algorithms have emphasized the need for direct methods to identify mRNA targets [64]. One such method used in flies is the identification of mRNAs present in Ago1 complexes purified from cell lines and embryos, since these IPs should be enriched for miRNA targets [67, 68]. Analogous methods have been used in other model systems [64, 69–71] which include a crosslinking step between protein and mRNA that facilitates the identification of miRNA-bound mRNA regions. More widespread use of these approaches in flies could identify in vivo miRNA targets and lead to considerably more accurate and powerful miRNA target prediction tools.
4.2 Strategies for Validating miRNA Targets
Targets identified from the approaches described above must be verified in vivo . A common and rapid method to determine whether a miRNA binding site is functional involves using cell culture -based reporter assay, in which the 3′UTR of the target gene of interest is placed downstream of a reporter gene. Reporters bearing mutated miRNA binding sites are used as negative control. MiRNAs are transfected, and the expression of the reporters was analyzed. Transgenic animals harboring such reporters are used to analyze predicted target 3′UTRs in vivo , and such verified reporters can also be used as the miRNA sensors described in Subheading 3.3.
The current gold standard to validate predicted miRNA targets relies on genetic manipulations. The genetic loss or gain of a miRNA should have a predictive effect on the expression of responsive mRNA targets, leading to their elevation or reduction respectively. Available resources to manipulate miRNA levels are described in Subheading 5 below. Furthermore, the functional relevance of individual miRNA–target relationships can be assessed with genetic interaction assays. Based on the assumption that miRNA phenotypes are a consequence of target overexpression, these assays usually test whether particular phenotypes are suppressed by the reduction of target expression due to heterozygosity or RNAi -mediated knock down. This approach has identified a number of functional miRNA/mRNA interactions, including between miR-8 and its targets atrophin [59], U-shaped [58], and components of the Toll pathway [72], miR-9a and its multiple targets dystroglycan [73], senseless [74], and dLMO [75], miR-14 and sugarbabe [61], miR-124 and its targets anachronism [21] and transformer [76], miR-184 and saxophone [77], miR-263a/b and head involution defective [78], miR-278 and expanded [22], miR-279 and its targets unpaired [60] and nerfin-1 [79], the miR-310/313 cluster and khc-73 [80] as well as the wingless pathway [81], and between the miR-100/let-7/miR-125 cluster and chronologically inappropriate morphogenesis [62] as well as a second target abrupt [82].
A very recently described approach employs genome editing to evaluate the functional and physiological relevance of particular miRNA–target relationships. The clustered regularly interspaced short palindromic repeat (CRISPR)/Cas9 system allows manipulation of sequences within endogenous genetic loci [83]. Using this system, Bassett et al. deleted predicted binding sites of the bantam miRNA within 3′UTR sequences of its previously established target, enabled [84]. Contrary to expectations, these manipulations did not lead to elevated Enabled expression in vivo or to known wing boundary phenotypes associated with elevated Enabled. While these results suggest that bantam may not regulate enabled during wing formation, it remains possible that the edited enabled 3′UTR retains cryptic or non-canonical bantam binding sites. Nevertheless, this approach will likely become a widely used assay to evaluate the physiological relevance of specific miRNA–target interactions, since it bypasses the potential pleiotropic effects resulting from miRNA knockout or overexpression.
5 In Vivo Functions
As with any other genes, the in vivo function of miRNAs is determined by analyzing the consequences of their overexpression and depletion in animals and cell lines . A series of recently described reagent collections greatly facilitate this type of analysis, and are summarized here.
5.1 Conditional Expression of microRNAs
Three groups have independently prepared collections of transgenic strains for the conditional expression of most fly miRNAs [24, 85, 86]. These three overlapping collections are based on the UAS /Gal4 system and contain individual or clusters of miRNAs under UAS control. The specifics of each collection vary, and are summarized in Table 1. Key differences between the collections include the design of transformation plasmid, the method of transgenesis, the location of the resulting transgenes, and number of individual miRNAs represented. Many miRNAs are included in all three collections, allowing the opportunity to easily verify results using independently generated but presumably equivalent reagents.
To date, these collections of UAS -miRNA transgenes have been used to screen for miRNAs whose elevated or ectopic expression lead to wing phenotypes and embryonic lethality [24, 85]. These types of analyses can be extended to additional tissues and developmental stages as well as to the cellular and molecular levels in order to verify predicted targets. Conditional expression of a genome-wide miRNA library has also been applied to cell culture in order to systematically identify miRNAs that regulate specific 3′UTRs [87]. This approach identified a set of miRNAs that regulate components of the hedgehog signaling pathway that were subsequently verified in intact animals, and can be used to identify miRNAs that regulate additional 3′UTRs of particular interest as well.
5.2 Genetic Depletion of microRNAs
MiRNA function has been inferred from the phenotypic analysis of mutants in core miRNA processing components as well as individual miRNAs or miRNA clusters. Genetic analysis of core components has usually focused on dicer, drosha, pasha, and argonaute, but is confounded by observations that these loci have nonoverlapping, miRNA-independent functions and consequently distinct phenotypes [26, 88]. Therefore, a clearer picture of the roles of miRNAs in particular biological processes can be obtained from individual mutants, although the creation of such mutants has historically been laborious and time-consuming. To date, mutations in 16 miRNA loci have been reported using homologous recombination and P-element excision, including miR-1, miR-7, miR-8, miR-9a, miR-9c, miR-11, miR-12/-283/-304, miR-34, miR-92b, miR-100/let-7/miR-125, miR-124, miR-263a/-263b, miR-278, miR-279/-996, miR-309/-3/-286/-4/-5/-6-1/-6-2/-6-3, miR-310/-311/-312/-313, miR-969, miR-989, bantam, and iab-4/-8 [19–22, 27, 39, 50, 59, 74, 77–80, 82, 89–98]. This collection of mutants disrupts a total of 33 miRNAs due to miRNA clustering, and rescuing transgenes are available to probe some of the individual members of such clusters.
The set of available miRNA mutants will expand considerably due to a large-scale effort by Stephen Cohen’s lab that has generated 77 new mutants by targeted homologous recombination (Stephen Cohen, personal communication). Together with previously reported mutants, the collection encompasses 95 mutant loci deleting 130 miRNAs that collectively account for the overwhelming majority (>99 %) of all D. melanogaster miRNA sequence reads. This resource will allow individual researchers to comprehensively evaluate the role of miRNAs in most developmental processes using standard techniques including mosaic analysis of cells and tissues of interest. Potential redundancy can be addressed via the combination of multiple mutants. Techniques will need to be developed to genetically deplete miRNAs in post-mitotic cells in order to examine their roles in differentiated cells , including during tissue remodeling and aging.
5.3 microRNA Sponges
miRNA sponges complement miRNA mutants for the analysis of loss-of-function phenotypes. Sponges are transgenically encoded transcripts that contain nucleotide repeats that are complementary to a particular miRNA. Conditional expression of these transcripts likely sequesters the targeted miRNA, interfering with its normal function. Sponges have been generated to 11 miRNAs to date, including miR-2b, -2c, -6, -7, -8, -9a, 13a, -13b, 92b, and -276 [85, 92, 96, 99]. Current sponge design includes 20 tandem repeats located within the 3′UTR of dsRed, which is used to monitor sponge expression, under UAS control. Case studies found that sponges for miR-7, -8, and 9a elicited similar though milder phenotypes as previously characterized genetic mutants [99]. Thus, current efforts to generate a comprehensive transgenic sponge library will facilitate functional analysis, allowing the use of specific Gal4 drivers to simultaneously knock down multiple miRNAs in specific populations of dividing or differentiated cells and tissues. However, since sponge transcript levels must be saturating, analysis is best performed with multiple insertions of the same sponge and in a background with reduced miRNA gene dosage. Even then, this approach may not effectively sequester the most abundant miRNAs, like miR-1. Furthermore, to eliminate concerns about false-positive and false-negative phenotypes, results with sponges will need to be confirmed with genetic mutants.
5.4 Other Antisense Techniques
In addition to transgenic sponges, other antisense technologies have been employed to disrupt fly miRNA function. For example, sequence-specific antisense oligonucleotides that block the function of individual miRNAs can be easily transfected into cultured cells in order to characterize cellular and molecular phenotypes [100]. An analogous approach has also been pioneered in intact adults that injected with cholesterol-modified antisense oligonucleotides to investigate the role of let-7 in post-mitotic neurons [101]. Key issues of working with such “antagomirs” include their effective and specific delivery as well as the occurrence of false-positive phenotypes due to off-target effects . The latter is of particular concern, given that embryonic phenotypes reported in an early study on antagomirs have not been shared by subsequently generated genetic mutants [102]. Thus, caution is merited when using antagomirs and any phenotypes identified from antisense injection should be verified in cells or tissues genetically depleted of the targeted miRNA.
6 Final Thoughts
The D. melanogaster model system provides an ever-increasing repertoire of resources and methods available to study miRNAs. With a manageable number of miRNA genes and mutations available in many of them, the role of miRNAs in complex biological processes can be comprehensively analyzed at molecular and cellular resolution. Thus, the fly model will likely continue to serve as a powerful tool to explore the biological roles of these fascinating molecules.
References
Ameres SL, Zamore PD (2013) Diversifying microRNA sequence and function. Nat Rev Mol Cell Biol 14(8):475–488
Ha M, Kim VN (2014) Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol 15(8):509–524
Kozomara A, Griffiths-Jones S (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res 42(Database issue):D68–D73
Sokol NS (2008) An overview of the identification, detection, and functional analysis of Drosophila microRNAs. Methods Mol Biol 420:319–334
Stark A et al (2007) Systematic discovery and characterization of fly microRNAs using 12 Drosophila genomes. Genome Res 17(12):1865–1879
Berezikov E et al (2010) Evolutionary flux of canonical microRNAs and mirtrons in Drosophila. Nat Genet 42(1):6–9, author reply 9–10
Berezikov E et al (2011) Deep annotation of Drosophila melanogaster microRNAs yields insights into their processing, modification, and emergence. Genome Res 21(2):203–215
Chung WJ et al (2011) Computational and experimental identification of mirtrons in Drosophila melanogaster and Caenorhabditis elegans. Genome Res 21(2):286–300
Ruby JG, Jan CH, Bartel DP (2007) Intronic microRNA precursors that bypass Drosha processing. Nature 448(7149):83–86
Ruby JG et al (2007) Evolution, biogenesis, expression, and target predictions of a substantially expanded set of Drosophila microRNAs. Genome Res 17(12):1850–1864
Wen J et al (2014) Diversity of miRNAs, siRNAs, and piRNAs across 25 Drosophila cell lines. Genome Res 24(7):1236–1250
Lau NC et al (2009) Abundant primary piRNAs, endo-siRNAs, and microRNAs in a Drosophila ovary cell line. Genome Res 19(10):1776–1785
Graveley BR et al (2011) The developmental transcriptome of Drosophila melanogaster. Nature 471(7339):473–479
Liu N et al (2011) The exoribonuclease Nibbler controls 3' end processing of microRNAs in Drosophila. Curr Biol 21(22):1888–1893
Han BW et al (2011) The 3'-to-5' exoribonuclease Nibbler shapes the 3' ends of microRNAs bound to Drosophila Argonaute1. Curr Biol 21(22):1878–1887
Chawla G, Sokol NS (2014) ADAR mediates differential expression of polycistronic microRNAs. Nucleic Acids Res 42(8):5245–5255
Okamura K et al (2013) Functional small RNAs are generated from select miRNA hairpin loops in flies and mammals. Genes Dev 27(7):778–792
Biemar F et al (2005) Spatial regulation of microRNA gene expression in the Drosophila embryo. Proc Natl Acad Sci U S A 102(44):15907–15911
Sokol NS, Ambros V (2005) Mesodermally expressed Drosophila microRNA-1 is regulated by Twist and is required in muscles during larval growth. Genes Dev 19(19):2343–2354
Kwon C et al (2005) MicroRNA1 influences cardiac differentiation in Drosophila and regulates Notch signaling. Proc Natl Acad Sci U S A 102(52):18986–18991
Weng R, Cohen SM (2012) Drosophila miR-124 regulates neuroblast proliferation through its target anachronism. Development 139(8):1427–1434
Teleman AA, Maitra S, Cohen SM (2006) Drosophila lacking microRNA miR-278 are defective in energy homeostasis. Genes Dev 20(4):417–422
Chawla G, Sokol NS (2012) Hormonal activation of let-7-C microRNAs via EcR is required for adult Drosophila melanogaster morphology and function. Development 139(10):1788–1797
Schertel C et al (2012) Functional characterization of Drosophila microRNAs by a novel in vivo library. Genetics 192(4):1543–1552
Pressman S et al (2012) A systematic genetic screen to dissect the MicroRNA pathway in Drosophila. G3 (Bethesda) 2(4):437–448
Luhur A et al (2014) Drosha-independent DGCR8/Pasha pathway regulates neuronal morphogenesis. Proc Natl Acad Sci U S A 111(4):1421–1426
Brennecke J et al (2003) bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila. Cell 113(1):25–36
Lai EC, Tam B, Rubin GM (2005) Pervasive regulation of Drosophila Notch target genes by GY-box-, Brd-box-, and K-box-class microRNAs. Genes Dev 19(9):1067–1080
Stark A et al (2003) Identification of Drosophila MicroRNA targets. PLoS Biol 1(3):E60
Forstemann K et al (2005) Normal microRNA maturation and germ-line stem cell maintenance requires Loquacious, a double-stranded RNA-binding domain protein. PLoS Biol 3(7):e236
Brennecke J et al (2005) Principles of microRNA-target recognition. PLoS Biol 3(3):e85
Yoo B et al (2014) Detection of miRNA expression in intact cells using activatable sensor oligonucleotides. Chem Biol 21(2):199–204
Toledano H et al (2012) Dual fluorescence detection of protein and RNA in Drosophila tissues. Nat Protoc 7(10):1808–1817
Aboobaker AA et al (2005) Drosophila microRNAs exhibit diverse spatial expression patterns during embryonic development. Proc Natl Acad Sci U S A 102(50):18017–18022
Kosman D et al (2004) Multiplex detection of RNA expression in Drosophila embryos. Science 305(5685):846
Stark A et al (2008) A single Hox locus in Drosophila produces functional microRNAs from opposite DNA strands. Genes Dev 22(1):8–13
Thomson JM et al (2006) Extensive post-transcriptional regulation of microRNAs and its implications for cancer. Genes Dev 20(16):2202–2207
Soni K et al (2013) miR-34 is maternally inherited in Drosophila melanogaster and Danio rerio. Nucleic Acids Res 41(8):4470–4480
Li X, Carthew RW (2005) A microRNA mediates EGF receptor signaling and promotes photoreceptor differentiation in the Drosophila eye. Cell 123(7):1267–1277
Kucherenko MM et al (2012) Steroid-induced microRNA let-7 acts as a spatio-temporal code for neuronal cell fate in the developing Drosophila brain. EMBO J 31(24):4511–4523
Lemons D, Pare A, McGinnis W (2012) Three Drosophila Hox complex microRNAs do not have major effects on expression of evolutionarily conserved Hox gene targets during embryogenesis. PLoS One 7(2):e31365
Marrone AK et al (2012) Dg-Dys-Syn1 signaling in Drosophila regulates the microRNA profile. BMC Cell Biol 13:26
Toledano H et al (2012) The let-7-Imp axis regulates ageing of the Drosophila testis stem-cell niche. Nature 485(7400):605–610
Laneve P, Giangrande A (2014) Enhanced northern blot detection of small RNA species in Drosophila melanogaster. J Vis Exp. doi:10.3791/51814
Okamura K et al (2007) The mirtron pathway generates microRNA-class regulatory RNAs in Drosophila. Cell 130(1):89–100
Varallyay E, Burgyan J, Havelda Z (2007) Detection of microRNAs by Northern blot analyses using LNA probes. Methods 43(2):140–145
Yang Y et al (2009) The bantam microRNA is associated with drosophila fragile X mental retardation protein and regulates the fate of germline stem cells. PLoS Genet 5(4):e1000444
Varghese J, Cohen SM (2007) microRNA miR-14 acts to modulate a positive autoregulatory loop controlling steroid hormone signaling in Drosophila. Genes Dev 21(18):2277–2282
Chen C et al (2005) Real-time quantification of microRNAs by stem-loop RT-PCR. Nucleic Acids Res 33(20):e179
Liu N et al (2012) The microRNA miR-34 modulates ageing and neurodegeneration in Drosophila. Nature 482(7386):519–523
Esslinger SM et al (2013) Drosophila miR-277 controls branched-chain amino acid catabolism and affects lifespan. RNA Biol 10(6):1042–1056
Daneshvar K et al (2013) MicroRNA miR-308 regulates dMyc through a negative feedback loop in Drosophila. Biol Open 2(1):1–9
Friedlander MR et al (2008) Discovering microRNAs from deep sequencing data using miRDeep. Nat Biotechnol 26(4):407–415
Pritchard CC, Cheng HH, Tewari M (2012) MicroRNA profiling: approaches and considerations. Nat Rev Genet 13(5):358–369
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12(2):87–98
Bentley DR et al (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456(7218):53–59
Bartel DP (2009) MicroRNAs: target recognition and regulatory functions. Cell 136(2):215–233
Hyun S et al (2009) Conserved MicroRNA miR-8/miR-200 and its target USH/FOG2 control growth by regulating PI3K. Cell 139(6):1096–1108
Karres JS et al (2007) The conserved microRNA miR-8 tunes atrophin levels to prevent neurodegeneration in Drosophila. Cell 131(1):136–145
Luo W, Sehgal A (2012) Regulation of circadian behavioral output via a MicroRNA-JAK/STAT circuit. Cell 148(4):765–779
Varghese J, Lim SF, Cohen SM (2010) Drosophila miR-14 regulates insulin production and metabolism through its target, sugarbabe. Genes Dev 24(24):2748–2753
Wu YC et al (2012) Let-7-complex microRNAs regulate the temporal identity of Drosophila mushroom body neurons via chinmo. Dev Cell 23(1):202–209
Chi SW, Hannon GJ, Darnell RB (2012) An alternative mode of microRNA target recognition. Nat Struct Mol Biol 19(3):321–327
Chi SW et al (2009) Argonaute HITS-CLIP decodes microRNA-mRNA interaction maps. Nature 460(7254):479–486
Smibert P et al (2012) Global patterns of tissue-specific alternative polyadenylation in Drosophila. Cell Rep 1(3):277–289
Rehmsmeier M et al (2004) Fast and effective prediction of microRNA/target duplexes. RNA 10(10):1507–1517
Easow G, Teleman AA, Cohen SM (2007) Isolation of microRNA targets by miRNP immunopurification. RNA 13(8):1198–1204
Kadener S et al (2009) A role for microRNAs in the Drosophila circadian clock. Genes Dev 23(18):2179–2191
Hafner M et al (2010) Transcriptome-wide identification of RNA-binding protein and microRNA target sites by PAR-CLIP. Cell 141(1):129–141
Zisoulis DG et al (2010) Comprehensive discovery of endogenous Argonaute binding sites in Caenorhabditis elegans. Nat Struct Mol Biol 17(2):173–179
Helwak A et al (2013) Mapping the human miRNA interactome by CLASH reveals frequent noncanonical binding. Cell 153(3):654–665
Lee GJ, Hyun S (2014) Multiple targets of the microRNA miR-8 contribute to immune homeostasis in Drosophila. Dev Comp Immunol 45(2):245–251
Yatsenko AS, Shcherbata HR (2014) Drosophila miR-9a targets the ECM receptor Dystroglycan to canalize myotendinous junction formation. Dev Cell 28(3):335–348
Li Y et al (2006) MicroRNA-9a ensures the precise specification of sensory organ precursors in Drosophila. Genes Dev 20(20):2793–2805
Bejarano F, Smibert P, Lai EC (2010) miR-9a prevents apoptosis during wing development by repressing Drosophila LIM-only. Dev Biol 338(1):63–73
Weng R et al (2013) miR-124 controls male reproductive success in Drosophila. Elife 2:e00640
Iovino N, Pane A, Gaul U (2009) miR-184 has multiple roles in Drosophila female germline development. Dev Cell 17(1):123–133
Hilgers V, Bushati N, Cohen SM (2010) Drosophila microRNAs 263a/b confer robustness during development by protecting nascent sense organs from apoptosis. PLoS Biol 8(6):e1000396
Cayirlioglu P et al (2008) Hybrid neurons in a microRNA mutant are putative evolutionary intermediates in insect CO2 sensory systems. Science 319(5867):1256–1260
Tsurudome K et al (2010) The Drosophila miR-310 cluster negatively regulates synaptic strength at the neuromuscular junction. Neuron 68(5):879–893
Pancratov R et al (2013) The miR-310/13 cluster antagonizes beta-catenin function in the regulation of germ and somatic cell differentiation in the Drosophila testis. Development 140(14):2904–2916
Caygill EE, Johnston LA (2008) Temporal regulation of metamorphic processes in Drosophila by the let-7 and miR-125 heterochronic microRNAs. Curr Biol 18(13):943–950
Bassett AR et al (2014) Understanding functional miRNA-target interactions in vivo by site-specific genome engineering. Nat Commun 5:4640
Becam I et al (2011) Notch-mediated repression of bantam miRNA contributes to boundary formation in the Drosophila wing. Development 138(17):3781–3789
Bejarano F et al (2012) A genome-wide transgenic resource for conditional expression of Drosophila microRNAs. Development 139(15):2821–2831
Szuplewski S et al (2012) MicroRNA transgene overexpression complements deficiency-based modifier screens in Drosophila. Genetics 190(2):617–626
Kim K, Vinayagam A, Perrimon N (2014) A rapid genome-wide microRNA screen identifies miR-14 as a modulator of Hedgehog signaling. Cell Rep 7(6):2066–2077
Yang JS, Lai EC (2011) Alternative miRNA biogenesis pathways and the interpretation of core miRNA pathway mutants. Mol Cell 43(6):892–903
Sokol NS et al (2008) Drosophila let-7 microRNA is required for remodeling of the neuromusculature during metamorphosis. Genes Dev 22(12):1591–1596
Bender W (2008) MicroRNAs in the Drosophila bithorax complex. Genes Dev 22(1):14–19
Bushati N et al (2008) Temporal reciprocity of miRNAs and their targets during the maternal-to-zygotic transition in Drosophila. Curr Biol 18(7):501–506
Chen Z et al (2012) miR-92b regulates Mef2 levels through a negative-feedback circuit during Drosophila muscle development. Development 139(19):3543–3552
Friggi-Grelin F, Lavenant-Staccini L, Therond P (2008) Control of antagonistic components of the hedgehog signaling pathway by microRNAs in Drosophila. Genetics 179(1):429–439
Kugler JM et al (2013) Maternal loss of miRNAs leads to increased variance in primordial germ cell numbers in Drosophila melanogaster. G3 (Bethesda) 3(9):1573–1576
Kugler JM et al (2013) miR-989 is required for border cell migration in the Drosophila ovary. PLoS One 8(7):e67075
Li W et al (2013) MicroRNA-276a functions in ellipsoid body and mushroom body neurons for naive and conditioned olfactory avoidance in Drosophila. J Neurosci 33(13):5821–5833
Sun K et al (2012) Neurophysiological defects and neuronal gene deregulation in Drosophila mir-124 mutants. PLoS Genet 8(2):e1002515
Xu P et al (2003) The Drosophila microRNA Mir-14 suppresses cell death and is required for normal fat metabolism. Curr Biol 13(9):790–795
Loya CM et al (2009) Transgenic microRNA inhibition with spatiotemporal specificity in intact organisms. Nat Methods 6(12):897–903
Horwich MD, Zamore PD (2008) Design and delivery of antisense oligonucleotides to block microRNA function in cultured Drosophila and human cells. Nat Protoc 3(10):1537–1549
Gehrke S et al (2010) Pathogenic LRRK2 negatively regulates microRNA-mediated translational repression. Nature 466(7306):637–641
Leaman D et al (2005) Antisense-mediated depletion reveals essential and specific functions of microRNAs in Drosophila development. Cell 121(7):1097–1108
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Chawla, G., Luhur, A., Sokol, N. (2016). Analysis of MicroRNA Function in Drosophila . In: Dahmann, C. (eds) Drosophila. Methods in Molecular Biology, vol 1478. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6371-3_4
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6371-3_4
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6369-0
Online ISBN: 978-1-4939-6371-3
eBook Packages: Springer Protocols