Abstract
Protein modification by the small ubiquitin-related modifier (SUMO) protein regulates numerous cellular pathways and mounting evidence reveals a critical role for SUMO in modulating gene expression. Dynamic sumoylation of transcription factors, chromatin-modifying enzymes, histones, and other chromatin-associated factors significantly affects the transcriptional status of the eukaryotic genome. Recent studies have employed high-throughput ChIP-Seq analyses to gain clues regarding the role of the SUMO pathway in regulating chromatin-based transactions. Indeed, the global distribution of SUMO across chromatin reveals an important function for SUMO in controlling transcription, particularly of genes involved in protein synthesis. These newly appreciated patterns of genome-wide sumoylation will inform more directed studies aimed at analyzing how the dynamics of gene expression are controlled by posttranslational SUMO modification.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 The SUMO Pathway
The small ubiquitin-related modifier (SUMO) protein is a conserved posttranslational modification that alters the binding, conformation, and/or localization of a substrate protein. Protein sumoylation is essential in most eukaryotes and regulates numerous cellular processes including mitochondrial dynamics, ribosome biogenesis, and DNA repair [1]. This review focuses on the recent and exciting body of work connecting protein SUMO modification to chromatin dynamics and transcriptional regulation. Particular emphasis will be placed on several novel high-throughput chromatin immunoprecipitation-DNA sequencing analyses (ChIP-Seq ) that have revealed the localization of SUMO across the genome and provide new insight into the role of SUMO in gene expression.
SUMO, like other members of the ubiquitin-like protein (UBL) family, covalently attaches to target proteins in a process similar to ubiquitin-protein conjugation (Fig. 1) [2]. SUMO is synthesized as an inactive precursor with a C-terminal peptide extension. A SUMO protease cleaves after a C-terminal-proximal Gly-Gly motif to form mature, conjugation-competent SUMO. In an ATP-dependent manner, the carboxyl-terminus of mature SUMO is activated by the heterodimeric SUMO-activating enzyme (E1), forming a high-energy thioester bond with the E1. SUMO is then transferred to the active-site cysteine of the E2 SUMO-conjugating enzyme followed by SUMO transfer to a lysine side chain on the target protein. Protein sumoylation is usually assisted by one of a small number of SUMO E3 ligases , which enhance conjugation specificity [2]. Target proteins can be sumoylated on a single lysine (monosumoylation), on multiple lysines with a single SUMO moiety (multisumoylation), or on one or more lysines with an extended SUMO chain (polysumoylation).
The SUMO pathway. The small ubiquitin-like modifier protein (SUMO) is first synthesized as an inactive precursor, which is processed by a SUMO protease to create mature, conjugatable SUMO. In an ATP-dependent manner, mature SUMO is activated and subsequently conjugated to the lysine side chain(s) of a substrate protein through the concerted action of E1, E2, and E3 enzymes
The budding yeast Saccharomyces cerevisiae expresses one form of SUMO (Smt3) , while most vertebrates have three active SUMO isoforms, SUMO-1, SUMO-2, and SUMO-3. The SUMO-2 and SUMO-3 proteins share ~97 % sequence identity and are sometimes referred to as SUMO-2/3. The yeast Smt3 protein has the highest sequence identity with human SUMO-1 (~48 %); however, while the Smt3 protein can form SUMO chains in vivo, SUMO-1 cannot [3–5]. Instead, SUMO-2 and SUMO-3 are the predominant chain formers in mammalian cells [6]. Protein substrates of sumoylation are often, but not always, modified on a SUMO consensus motif, which is a stretch of four amino acids with the sequence ψ-K-X-D/E (where ψ is a hydrophobic amino acid and X is any amino acid) [7, 8]. Additionally, other posttranslational modifications, notably phosphorylation, can also stimulate sumoylation [9–11].
Sumoylation is a dynamic modification. SUMO proteases specifically cleave the bond between SUMO and substrate. There are three known classes of SUMO proteases in vertebrates, the SENP/Ulp, DESI, and USPL1 families, with the SENP family being the largest—seven members in humans—and best characterized [12]. In yeast , where the SUMO proteases were first identified, only SENP/Ulp-class SUMO proteases, Ulp1 and Ulp2 , have been found to date [13, 14]. As with SUMO conjugation, desumoylation of proteins is highly regulated and plays a crucial role in many cellular pathways [15, 16].
The consequences of protein sumoylation are numerous and include changes in protein localization, altered protein conformation, and either enhanced or impaired protein-protein interactions . Frequently, the assembly and dynamics of large protein complexes are mediated through the interaction of sumoylated proteins with SUMO-interacting motifs (SIMs) of other proteins within the complex. SIMs interact with the β2 strand of SUMO and most are characterized by a core of 3–4 hydrophobic residues (typically Val or Ile) that is often flanked on one or the other side by acidic amino acids (Asp, Glu, or phosphorylated Ser or Thr) [17–19]. Thus, SUMO often acts as a molecular adhesive, bringing protein complexes together in a regulated fashion, as seen with the numerous proteins involved in DNA repair by homologous recombination [20]. Consequently, assessing the effects of protein sumoylation is often challenging because removal of one sumoylation site within a protein complex rarely produces an observable phenotype. Therefore, identifying most or all sites of sumoylation within a protein assembly is often required to understand the precise role of SUMO regulation for a particular cellular process.
2 SUMO and Transcription: The Example of the Yeast Tup1-Ssn6 Corepressor
Sumoylation contributes broadly to the regulation of gene expression, with many studies finding that SUMO inhibits transcription [21]. However, in budding yeast sumoylation is required for efficient RNA polymerase II recruitment to constitutively expressed genes, as well as for coordinating the proper activation and inactivation kinetics of several inducible genes [22, 23]. Furthermore, several genome-wide ChIP studies have shown that SUMO is present at the promoters of many constitutive genes, including ribosomal protein genes, in both yeast and humans [24–26]. SUMO modification of proteins involved in transcription has been reviewed extensively [27–30], and thus we will focus on recent insights into the consequences of sumoylation of the yeast general transcriptional corepressors Tup1 and Ssn6 as an illustration of how these modifications alter gene expression.
In S. cerevisiae, transcription of the galactose-inducible GAL genes is tightly regulated by carbon source [31]. Full repression of the GAL genes, as occurs when cells are grown in glucose, is mediated by the Mig1 transcriptional repressor along with the corepressors Tup1 and Ssn6 [32, 33]. When cells are shifted from glucose to galactose growth media, the GAL genes are slowly derepressed before full activation. Derepression of the GAL1 gene (glucose to galactose) requires the SUMO protease Ulp1 ; however, Ulp1 is not required for GAL1 activation from an inactive but not fully repressed state (for example, by switching from raffinose to galactose) [34]. When Ulp1 is untethered from the nuclear pore complex (NPC), where it normally concentrates, or when the catalytic domain of Ulp1 is artificially tethered to the GAL1 locus, GAL1 derepression kinetics are enhanced compared to wild-type yeast [34]. The corepressors Ssn6 and Tup1 are both sumoylated in vivo and are Ulp1 substrates. Mutation of Ssn6 SUMO consensus attachment sites results in faster GAL1 derepression, indicating that Ssn6 desumoylation is likely required for proper GAL1 up-regulation upon a shift from glucose to galactose. The model proposed from this study is that GAL1 gene activation involves recruitment of the repressed GAL1 locus to the NPC, and consequent Ssn6 desumoylation by NPC-localized Ulp1, enabling subsequent recruitment of transcriptional activators through an undetermined molecular mechanism. This mode of regulation was also observed for the glucose-repressed gene HXK1, indicating a potentially general role for Ulp1 -mediated desumoylation in inducible gene activation.
Tup1 sumoylation also represses gene expression, particularly in response to various stress conditions [35]. Tup1 is normally recruited to the promoters of the inducible genes ARG1 and CPA2 following amino acid starvation, as revealed by time-course ChIP analysis. Using a “SUMOless” mutant of Tup1, Ng et al. [35] found that initial Tup1 recruitment to the promoter regions is not hampered following amino acid starvation. However, the maintenance of Tup1 association with promoter regions is reduced with the “SUMOless” mutant, leading to an extended period of RNA polymerase II occupancy at the promoters and a consequent increase in ARG1 and CPA2 mRNA levels. These results suggest that Tup1 sumoylation helps to deactivate inducible genes following an initial period of stress-induced transcription. Taken together, these studies highlight an important function for SUMO in fine-tuning inducible gene expression.
3 SUMO Localization Across the Genome
To gain a deeper understanding of SUMO function in transcription and chromatin dynamics, it is often essential to dissect the consequences of sumoylation on one particular protein substrate or group of proteins [20]. In recent work, however, researchers have also zoomed out to analyze the broad distribution of SUMO across the genome, with the aim of determining a more global and general role for SUMO modification in gene expression. Using chromatin immunoprecipitation followed by high-throughput DNA sequencing (ChIP-Seq ), several groups have monitored the pattern of SUMO binding to chromatin in both mammalian and yeast cells (Fig. 2) and under normal growth conditions and during periods of stress [24–26, 36–38]. Interestingly, these studies reveal SUMO enrichment in areas of active gene transcription, particularly within the upstream promoter region of many constitutively expressed genes. Furthermore, sumoylation appears to control the expression of many genes involved in protein synthesis, thereby linking SUMO with the transcriptional regulation of cell growth and proliferation.
SUMO localization across the genome. SUMO ChIP-Seq studies revealed the pattern of SUMO across the yeast and mammalian genomes. In Saccharomyces cerevisiae (left panel ), sumoylation of the transcription factor Rap1 localizes it to the promoters of ribosomal protein (RP) genes. Sumoylated Rap1 recruits RNA polymerase II to these gene sites, and stimulates their transcription. Similarly, in mammalian cells (right panel ), SUMO-1 and SUMO-2/3 are primarily concentrated at the promoter regions of RP genes. However, SUMO localization at these regions has been shown to correlate with both activation and repression of RP gene transcription
4 SUMO Across the Mammalian Genome
In the first ChIP-Seq study of SUMO localization along mammalian chromosomes , Liu et al. analyzed the pattern of SUMO-1 localization on HeLa cell genomic DNA as a function of cell-cycle stage [26]. The genome-wide distribution of SUMO-modified substrates was assessed by next-generation sequencing of the protein-bound DNA fragments. Contrary to studies showing association of SUMO-1 with repressive elements, the authors found SUMO-1 to be enriched at promoters of active genes, particularly during interphase (namely G1 through late S phase). This association of SUMO-1 with active gene promoters decreased during mitosis , the least transcriptionally active cell-cycle stage. The promoter regions marked by SUMO-1 included many housekeeping genes, particularly ribosomal protein (RP) genes and other factors involved in translation. Further correlating SUMO with active transcription, approximately 70 % of promoters marked with SUMO-1 were enriched for histone H3 trimethylated on lysine-4 (H3K4me3 ), an active transcriptional mark, while only 9 % of SUMO-1 peaks overlapped with the repressive H3K27me3 mark.
To confirm a positive role for SUMO-1 in gene activation, Liu et al. [26] depleted SUMO-1 from HeLa cells using siRNA-mediated depletion of the SUMO-1 mRNA, and changes in gene expression were monitored by RNA-Seq. In total, 357 genes were differentially expressed compared to wild-type cells, with 199 of these genes being down-regulated in the SUMO-1-depleted cells. Gene Ontology (GO) analysis of these down-regulated genes showed a significant enrichment in genes involved in translation, thus indicating a positive role for SUMO-1 in transcriptional activation of protein synthesis genes. On the other hand, some transcript levels increased following SUMO-1 depletion; therefore, these transcripts are normally repressed by SUMO-1 in some way. It is possible that the effects of SUMO-1 siRNA on the transcription of some genes are indirect, or that SUMO-1 affects other steps in mRNA production. To further complicate matters, mRNA levels of several ribosomal protein genes, as determined by RT-qPCR (reverse transcription-quantitative polymerase chain reaction) , were differentially regulated in cells depleted of SUMO-1 or Ubc9 , the E2 SUMO-conjugating enzyme. Several transcripts down-regulated in SUMO-1 knockdown cells were instead up-regulated in Ubc9-depleted cells. It is possible that SUMO-2/3, which is also conjugated to proteins by Ubc9, antagonistically affects transcription of these particular ribosomal protein genes. These findings imply a more intricate role for SUMO-1 modification of transcription factors and chromatin-associated proteins, with some specific genes being activated by SUMO while others are repressed.
A mass spectrometry analysis for SUMO-1-modified proteins from S-phase HeLa cell lysates identified the DNA-binding protein scaffold-associated factor B1/2 (SAFB1/2) , a multifunctional protein that interacts with both RNA polymerase II and RNA-processing proteins [36]. Knockdown of SAFB1/2 in HeLa cells reduced SUMO-1 occupancy of the promoter regions of RP genes, suggesting that SAFB1/2 or SAFB1/2 interactors are the major proteins sumoylated at these promoters. Additionally, RNA polymerase II occupancy at RP gene promoters was diminished in SAFB siRNA-treated cells. Liu et al. [36] also monitored pre-mRNA splicing of two RP genes, RPL26 and RPL27a, which are regulated by SAFB1/2. Neither SUMO-1 nor SAFB1/2 depletion affected unspliced primary transcript levels of RPL26 and RPL27a; however, their knockdown did reduce spliced mRNA levels, revealing a novel effect of SAFB1/2 sumoylation on ribosomal protein gene expression, potentially through a dual—and possibly coupled—function in RNA polymerase II recruitment and pre-mRNA splicing.
Another recent report has also connected SUMO modification of chromatin factors with the regulation of RP gene transcription [24]. Neyret-Kahn et al. used ChIP-Seq to perform an in-depth analysis of SUMO-1, SUMO-2/3, Ubc9 , and PIASY (a mammalian SUMO E3 ligase ) binding to chromatin in proliferating human fibroblasts. As with the previous study, it was found that SUMO-1, and also SUMO-2/3, localizes to the transcription start site of many active gene promoters. SUMO-1 and SUMO-2/3 have similar association patterns (~two-thirds overlap of binding peaks); however there are genomic regions that only bind to one or the other paralog , indicating potentially unique roles for SUMO-1 and SUMO-2/3 in gene expression regulation. A strong correlation in genomic localization was observed in fibroblasts among SUMO, RNA polymerase II, and H3K4me3 , paralleling the SUMO-1 patterns observed in HeLa cells [26].
Fibroblast mRNA-Seq results revealed that 67 % of the genes marked by SUMO-1 or SUMO-2/3 were significantly expressed. Moreover, occupancy ranking of genes marked by SUMO-1 and SUMO-2/3 showed enrichment for histone genes, as well as genes involved with translation (e.g., RP genes), RNA Pol III-transcribed tRNA genes, and RNA Pol I-transcribed rRNA genes. These results again suggest an integral function for SUMO modifications at chromatin sites that regulate expression of genes important for protein synthesis.
Based on work in fibroblasts and HeLa cells, the precise roles for SUMO in regulating protein synthesis genes might well depend on cell type, as SUMO has been found to both positively and negatively regulate expression of specific RP genes. Moreover, the expression levels of several RNA Pol III transcripts (RNA5S, RN7SL1, and tRNA-Tyr), rRNA, and ribosomal protein genes RPL26 and RPS14 were modestly increased in both UBC9 and SUMO (SUMO-1 + SUMO-2/3) knockdown fibroblasts [24]. These results suggest a repressive role for sumoylation in the regulation of certain genes involved in translation, contrary to the results observed in HeLa cells [26]. Interpretation of the results in HeLa cells is confounded by the observation that mRNA levels of several ribosomal protein genes were higher in Ubc9 -depleted cells but lower in SUMO-1-depleted cells. It is possible that either all the SUMO paralogs together or the E2-conjugating enzyme Ubc9 must be knocked down in order to observe loss of the repressive effects of sumoylation on RP gene expression. Nevertheless, in their follow-up paper, Liu et al. [36] identified the chromatin scaffold protein SAFB1 as a sumoylated factor whose sumoylation correlates with enhanced pol II recruitment and pre-mRNA splicing of several RP genes. Thus, it is likely that SUMO-mediated control of gene expression is complex and that SUMO-dependent expression changes will depend on cell type, growth conditions, or other experimental factors.
5 SUMO Across the Yeast Genome
The global localization of SUMO (Smt3) on S. cerevisiae chromatin is similar to its localization in human cell lines in that SUMO clusters near the transcription start sites of many RP genes and tRNA genes [25]. Of the 395 unique SUMO ChIP-Seq peaks, 246 were at RNAPIII-transcribed tRNA genes, 110 at RP genes (out of 138 RP genes), 12 at non-ribosomal protein-coding genes, and 27 at genes for noncoding RNAs. A genome-wide RNA-Seq analysis of a temperature-sensitive ubc9-1 mutant, which is severely impaired for SUMO ligation, revealed no effect on global transcription, but showed that expression of nearly all RP genes was decreased [25]. Thus, Ubc9 , presumably through SUMO conjugation to specific gene-proximal chromatin components, stimulates yeast RP gene expression.
The SUMO-binding sites within the RP gene promoters were found to be similar to the consensus DNA-binding motif of the transcription factor Rap1 [25]. Rap1 ChIP-Seq analysis revealed strong colocalization of Rap1 and SUMO at RP gene promoters. Rap1 is a SUMO target, and blocking Rap1 sumoylation by mutating nine of its lysines to arginine, Rap1-K9R, reduced SUMO enrichment at RP gene promoters (but not Rap1 binding) and resulted in reduced RP gene expression. RNA polymerase II and TFIID (an RNA polymerase II preinitiation complex factor) binding to the RP gene promoters was reduced in ubc9-1 cells, suggesting that Rap1 sumoylation may be required for recruitment of RNA polymerase II and TFIID to RP gene promoters. Indeed, cells expressing the “SUMOless” Rap1-K9R have decreased binding of RNA polymerase II and TFIID at these promoters.
This study by Chymkowitch et al. [25] highlights a novel role for SUMO in regulating RP gene expression in yeast through modification of the conserved Rap1 transcription factor , thereby connecting the SUMO pathway to a regulatory mechanism of cell proliferation. It also opens up many avenues of study regarding SUMO dynamics and the control of transcription, including the role of SUMO in tRNA transcription and the molecular mechanism of TFIID recruitment to RP gene promoters by sumoylated Rap1.
6 SUMO-Chromatin Interactions During Heat Shock
Global sumoylation increases dramatically following heat shock, and targeted proteins include the heat-shock factors 1 and 2 (HSF1 and HSF2), other transcription factors , and numerous chromatin-associated proteins [39–49]. Recent genomic studies by both Niskanen et al. and Seifert et al. investigated the changes in chromatin-bound SUMO in response to heat shock [37, 38]. Both studies found that global SUMO-2/3 patterns change drastically across the mammalian genome in response to heat shock. During the stress, SUMO-2/3 accumulated at the promoter regions of actively transcribed genes. However, SUMO-2/3 does not simply stimulate transcription at these gene regions during heat shock. In leukemia and prostate cancer cells, knockdown of Ubc9 or PIAS1 by RNA interference, which causes a general drop in protein sumoylation, led to increased expression of heat-shock genes with normally SUMO-2/3-enriched promoters [37], suggesting that SUMO-2/3 ligation normally represses heat shock-induced gene expression, possibly to prevent hyperactivation of heat-shock genes during acute temperature stress. Contrary to the above study, Seifert et al. [38] found that SUMO-2/3 accumulation at actively transcribed regions during heat shock did not alter gene expression at these sites, but did likely influence the stability of protein complexes bound to the chromatin.
These genome-wide ChIP-Seq studies revealed the dynamic nature of sumoylation across the chromatin landscape in response to heat shock. The mechanism of regulating this massive shift in sumoylation likely involves the coordination of multiple signals and factors. For example, SUMO proteases might need to remove SUMO from chromatin-bound factors that are no longer sumoylated during heat shock, although it is possible that the sumoylated protein conjugate is removed intact by other mechanisms. The mammalian SUMO protease SENP6 is recruited to transcriptionally active DNA regions during heat shock [38]. Further work aimed at studying factors that affect the dynamics of SUMO on chromatin in response to different stresses will provide greater insight into the function of SUMO in regulating gene expression during heat shock and other changes in the cellular environment.
7 SUMO and Histones
Many chromatin-associated proteins are sumoylated, as discussed above. In addition to transcription factors , chromatin remodelers, and other chromatin-modifying enzymes, the central components of chromatin, the histone proteins, are also sumoylated. In budding yeast , all four core histone proteins and the histone variant H2A.Z can be detected in SUMO-modified forms [50, 51], while only sumoylated histones H3 and H4 have been identified in mammalian cells [46, 52]. Sumoylated histones have been discovered in a diverse range of organisms, including plants [53] and apicomplexan parasites (Plasmodium falciparum) [54], implicating histone-SUMO conjugation as a conserved and significant chromatin posttranslational modification. Thus, the already complicated and diverse histone code is now revealed to be even more complex, with a vast number and combination of possible histone posttranslational modifications. Understanding the interplay of these varied modifications will be necessary for elucidating the intricacies of gene regulation by histone alteration.
8 Histone Sumoylation Regulates Transcription
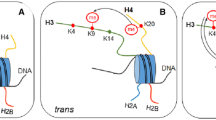
The four core histone proteins, H2A , H2B, H3 , and H4, are subject to various posttranslational modifications (PTMs), including acetylation, methylation, and ubiquitylation [55]. SUMO modifies lysine side chains of histones. First identified in mammalian cells in 2003, histone sumoylation appears to play a repressive role in transcription [52]. Histone H4 sumoylation in human cells recruits the histone deacetylase HDAC1 and heterochromatin protein 1 (HP1) to DNA, two factors involved in repressing transcription and maintaining silenced regions of the genome. All four yeast histone proteins were found to be sumoylated in vivo, and as seen in mammalian cells, SUMO-modified histones dampen transcriptional activity [50]. One possible mode of gene repression through histone sumoylation is by opposing activating histone modifications (i.e., acetylation). Indeed, yeast histone H2B sumoylation followed an inverse trend as compared to acetylation at the GAL1 locus upon activation. Since only a small fraction of bulk histone H2B is sumoylated, this repressive mechanism would require either a localized buildup of SUMO-H2B or SUMO-induced changes to H2B that persist after SUMO deconjugation that inhibits acetylation. More generally, histone sumoylation may have multiple modes of transcriptional regulation, including recruiting transcriptional repressors to gene promoters and blocking activating histone modifications.
Interestingly, a recent report by Hendriks et al. suggests that histone acetylation in human (HeLa) cells may stimulate sumoylation at a nearby lysine residue [46]. Human histone H3 was found by mass spectrometry to be simultaneously modified by SUMO, at Lys19, and by an acetyl group, at Lys24. When HeLa cells were treated with the histone deacetylase inhibitor trichostatin A, histone H3 sumoylation also increased. Conversely, histone acetyltransferase inhibition (curcumin) led to a corresponding decrease in H3 sumoylation. These results highlight a new example of cross talk between two different histone modifications, the function of which remains to be determined.
A recent in vitro study suggests a role for histone sumoylation in modulating chromatin structure and long-range chromatin interactions [56]. Dhall et al. created a disulfide-linked SUMO-3-histone H4 (at Lys-12) conjugate, showing that the SUMO-H4 readily incorporates into histone octamers and 12-mer nucleosome arrays. However, histone H4 sumoylation thwarted nucleosome compaction. Förster resonance energy transfer (FRET) measurement of internucleosomal interactions showed that histone H4 sumoylation reduces the affinity between two adjacent mononucleosomes. The basic N-terminal tail of histone H4 is important for establishing chromatin compaction, and accordingly, modification of residues within this region leads to changes in chromatin structure and organization [57]. Whether the mechanism of chromatin rearrangement by H4 Lys-12 sumoylation holds true in vivo remains to be determined; however, these findings suggest an additional potential mode of chromatin regulation mediated by histone sumoylation.
Despite these recent studies examining the function of histone sumoylation, our understanding of histone sumoylation remains limited. For example, the distribution of site-specific histone sumoylation across the genome has not been resolved. Furthermore, the dynamics of SUMO modification of histone proteins, including the possible regulated desumoylation by SUMO proteases, have yet to be worked out.
9 SUMO and Chromatin-Modifying Enzymes
SUMO also functions indirectly to modulate other histone PTMs by altering the activity of chromatin-modifying enzymes. Sumoylation of chromatin modifiers has varying consequences for the function and stability of the targeted enzyme. For example, sumoylation can enhance activity, as observed with SUMO-1 modification of the histone deacetylase HDAC4 [58]. Conversely, sumoylation of the histone lysine methyltransferase JARID1B/KDM5B leads to its RNF4 -mediated degradation , both during cell cycle progression and in response to DNA damage [59, 60]. RNF4 is a SUMO-targeted ubiquitin ligase (STUbL).
Frequently, SUMO modification of a transcription factor leads to recruitment of histone deacetylases, and thus repression of gene expression [61–63]. Sumoylation of the transcription factor Elk-1 recruits HDAC-2 to chromatin, which in turn reduces histone acetylation and consequently dampens transcription [61]. Intriguingly, SUMO modification both enhances and inhibits the interaction between chromatin factors and the histone methyltransferase SETDB1, depending on the identity of the modified protein [64, 65]. Conjugation of SUMO-1 to methyl-CpG-binding domain protein 1 (MBD1) reduced binding to SETDB1 and hence failed to repress transcription through histone methylation [64]. Conversely, sumoylation of the KAP1 corepressor recruited SETDB1 to chromatin, stimulated SETDB1 methyltransferase activity, and decreased gene expression [65]. SUMO modification appears to alter chromatin status through multiple mechanisms, and often the molecular consequences of these changes are target and context dependent.
A recent report by Nayak et al. has identified a role for dynamic desumoylation in the regulation of HOX gene expression, as controlled by the MLL1/MLL2 histone methyltransferase complexes [66]. HOX genes encode homeobox (HOX)-containing transcription factors crucial for vertebrate development. Transcriptional activation of these genes is tightly regulated. RbBP5, one of the four regulatory subunits of the MLL1/MLL2 complexes, is desumoylated by the SUMO protease SENP3 . Removal of SUMO-2 from RbBP5 in turn recruits the MLL components Ash2L and menin to a subgroup of HOX genes. The fully assembled MLL1/MLL2 complexes trimethylate H3K4 and recruit RNA polymerase II to promoter regions of the HOX genes, thereby turning on gene expression. These findings link SENP3 and the SUMO pathway to transcription-mediated modulation of a key developmental program.
10 Conclusions and Future Directions
The SUMO pathway is intimately linked to the control of gene expression. Covalent SUMO modification of transcription factors , chromatin-remodeling enzymes, and various other chromatin-related factors regulates transcription, not only in a repressive manner as originally observed (Fig. 3), but at times also in a stimulatory capacity. Novel ChIP-Seq investigations have shed light on the distribution of SUMO across the genomes of both budding yeast and mammalian cells. SUMO was found primarily at the promoter region of actively transcribed genes, most notably those of ribosomal protein genes. However, whether SUMO functions to activate or repress transcription at these genomic regions remains to be conclusively established, and the answer may depend on cell type, organism, and/or environmental conditions.
Dynamic SUMO modification regulates transcription. (a) Desumoylation of the yeast transcriptional co-repressor Ssn6 by the SUMO protease Ulp1 results in activation of the GAL1 gene following a shift to galactose-containing medium. (b) Sumoylation of histone core proteins, namely H2B and H4, represses transcription of yeast genes. One possible mechanism for transcriptional repression by histone-SUMO modification is through blocking sites of histone acetylation, an activating histone mark catalyzed by histone acetyltransferases (HATs). (c) Activation of mammalian histone methyltransferase complexes MLL1/MML2 requires desumoylation of the RbBP5 subunit by SENP3. Once activated and fully assembled, MLL1/MLL2 methylate lysine-4 of histone H3 that is present at HOX gene promoters, which leads to RNA polymerase II recruitment and transcriptional activation
Nonetheless, the results of these genomic studies have uncovered an unanticipated role for SUMO in regulating genes involved in protein synthesis and have implicated the SUMO pathway in controlling cell growth and the nutrient response. Several chromatin-binding proteins were found to be sumoylated when localized to ribosomal protein gene promoters, namely SAFB1/2 in mammalian cells and Rap1 in yeast . It will be important to determine if there are additional specific factors sumoylated at RP promoters in order to understand the exact role of SUMO at these sites.
The dynamic nature of SUMO-protein modification, through the action of SUMO proteases, allows for fine-tuning of cellular pathways controlled by sumoylation. The role of SUMO proteases in regulating expression of RP genes and other actively transcribed genes under stress conditions has yet to be studied. There are likely functions for SUMO proteases at these specific chromatin regions, particularly when the stimulatory or inhibitory effect of SUMO on transcription is no longer required.
References
Flotho A, Melchior F (2013) Sumoylation: a regulatory protein modification in health and disease. Annu Rev Biochem 82:357–385
Gareau JR, Lima CD (2010) The SUMO pathway: emerging mechanisms that shape specificity, conjugation and recognition. Nat Rev Cell Biol 11(12):861–871
Bylebyl GR, Belichenko I, Johnson ES (2003) The SUMO isopeptidase Ulp2 prevents accumulation of SUMO chains in yeast. J Biol Chem 278(45):44113–44120
Johnson ES, Gupta AA (2001) An E3-like factor that promotes SUMO conjugation to the yeast septins. Cell 106(6):735–744
Bencsath KP, Podgorski MS, Pagala VR, Slaughter CA, Schulman BA (2002) Identification of a multifunctional binding site on Ubc9p required for Smt3p conjugation. J Biol Chem 277(49):47938–47945
Tatham MH et al (2001) Polymeric chains of SUMO-2 and SUMO-3 are conjugated to protein substrates by SAE1/SAE2 and Ubc9. J Biol Chem 276(38):35368–35374
Minty A, Dumont X, Kaghad M, Caput D (2000) Covalent modification of p73alpha by SUMO-1. Two-hybrid screening with p73 identifies novel SUMO-1-interacting proteins and a SUMO-1 interaction motif. J Biol Chem 275(46):36316–36323
Rodriguez MS, Dargemont C, Hay RT (2001) SUMO-1 conjugation in vivo requires both a consensus modification motif and nuclear targeting. J Biol Chem 276(16):12654–12659
Hietakangas V et al (2006) PDSM, a motif for phosphorylation-dependent SUMO modification. Proc Natl Acad Sci U S A 103(1):45–50
Matic I et al (2010) Site-specific identification of SUMO-2 targets in cells reveals an inverted SUMOylation motif and a hydrophobic cluster SUMOylation motif. Mol Cell 39(4):641–652
Mohideen F et al (2009) A molecular basis for phosphorylation-dependent SUMO conjugation by the E2 UBC9. Nat Struct Mol Biol 16(9):945–952
Hickey CM, Wilson NR, Hochstrasser M (2012) Function and regulation of SUMO proteases. Nat Rev Mol Cell Biol 13(12):755–766
Li SJ, Hochstrasser M (1999) A new protease required for cell-cycle progression in yeast. Nature 398(6724):246–251
Li SJ, Hochstrasser M (2000) The yeast ULP2 (SMT4) gene encodes a novel protease specific for the ubiquitin-like Smt3 protein. Mol Cell Biol 20(7):2367–2377
Nayak A, Muller S (2014) SUMO-specific proteases/isopeptidases: SENPs and beyond. Genome Biol 15(7):422
Mukhopadhyay D, Dasso M (2007) Modification in reverse: the SUMO proteases. Trends Biochem Sci 32(6):286–295
Kerscher O (2007) SUMO junction-what's your function? New insights through SUMO-interacting motifs. EMBO Rep 8(6):550–555
Song J, Durrin LK, Wilkinson TA, Krontiris TG, Chen Y (2004) Identification of a SUMO-binding motif that recognizes SUMO-modified proteins. Proc Natl Acad Sci U S A 101(40):14373–14378
Hecker CM, Rabiller M, Haglund K, Bayer P, Dikic I (2006) Specification of SUMO1- and SUMO2-interacting motifs. J Biol Chem 281(23):16117–16127
Psakhye I, Jentsch S (2012) Protein group modification and synergy in the SUMO pathway as exemplified in DNA repair. Cell 151(4):807–820
Gill G (2005) Something about SUMO inhibits transcription. Curr Opin Genet Dev 15(5):536–541
Rosonina E, Duncan SM, Manley JL (2010) SUMO functions in constitutive transcription and during activation of inducible genes in yeast. Genes Dev 24(12):1242–1252
Rosonina E, Duncan SM, Manley JL (2012) Sumoylation of transcription factor Gcn4 facilitates its Srb10-mediated clearance from promoters in yeast. Genes Dev 26(4):350–355
Neyret-Kahn H et al (2013) Sumoylation at chromatin governs coordinated repression of a transcriptional program essential for cell growth and proliferation. Genome Res 23(10):1563–1579
Chymkowitch P et al (2015) Sumoylation of Rap1 mediates the recruitment of TFIID to promote transcription of ribosomal protein genes. Genome Res 25(6):897–906
Liu HW et al (2012) Chromatin modification by SUMO-1 stimulates the promoters of translation machinery genes. Nucleic Acids Res 40(20):10172–10186
Cubenas-Potts C, Matunis MJ (2013) SUMO: a multifaceted modifier of chromatin structure and function. Dev Cell 24(1):1–12
Raman N, Nayak A, Muller S (2013) The SUMO system: a master organizer of nuclear protein assemblies. Chromosoma 122(6):475–485
Garcia-Dominguez M, Reyes JC (2009) SUMO association with repressor complexes, emerging routes for transcriptional control. Biochim Biophys Acta 1789(6–8):451–459
Lyst MJ, Stancheva I (2007) A role for SUMO modification in transcriptional repression and activation. Biochem Soc Trans 35(Pt 6):1389–1392
Johnston M (1999) Feasting, fasting and fermenting. Glucose sensing in yeast and other cells. Trends Genet 15(1):29–33
Frolova E, Johnston M, Majors J (1999) Binding of the glucose-dependent Mig1p repressor to the GAL1 and GAL4 promoters in vivo: regulationby glucose and chromatin structure. Nucleic Acids Res 27(5):1350–1358
Treitel MA, Carlson M (1995) Repression by SSN6-TUP1 is directed by MIG1, a repressor/activator protein. Proc Natl Acad Sci U S A 92(8):3132–3136
Texari L et al (2013) The nuclear pore regulates GAL1 gene transcription by controlling the localization of the SUMO protease Ulp1. Mol Cell 51(6):807–818
Ng CH et al (2015) Sumoylation controls the timing of Tup1-mediated transcriptional deactivation. Nat Commun 6:6610
Liu HW, Banerjee T, Guan X, Freitas MA, Parvin JD (2015) The chromatin scaffold protein SAFB1 localizes SUMO-1 to the promoters of ribosomal protein genes to facilitate transcription initiation and splicing. Nucleic Acids Res 43(7):3605–3613
Niskanen EA et al (2015) Global SUMOylation on active chromatin is an acute heat stress response restricting transcription. Genome Biol 16:153
Seifert A, Schofield P, Barton GJ, Hay RT (2015) Proteotoxic stress reprograms the chromatin landscape of SUMO modification. Sci Signal 8(384):rs7
Saitoh H, Hinchey J (2000) Functional heterogeneity of small ubiquitin-related protein modifiers SUMO-1 versus SUMO-2/3. J Biol Chem 275(9):6252–6258
Tatham MH, Matic I, Mann M, Hay RT (2011) Comparative proteomic analysis identifies a role for SUMO in protein quality control. Sci Signal 4(178):rs4
Golebiowski F et al (2009) System-wide changes to SUMO modifications in response to heat shock. Science signaling 2(72):ra24
Miller MJ et al (2013) Quantitative proteomics reveals factors regulating RNA biology as dynamic targets of stress-induced SUMOylation in Arabidopsis. Mol Cell Proteom 12(2):449–463
Kurepa J et al (2003) The small ubiquitin-like modifier (SUMO) protein modification system in Arabidopsis. Accumulation of SUMO1 and -2 conjugates is increased by stress. J Biol Chem 278(9):6862–6872
Bruderer R et al (2011) Purification and identification of endogenous polySUMO conjugates. EMBO Rep 12(2):142–148
Lewicki MC, Srikumar T, Johnson E, Raught B (2015) The S. cerevisiae SUMO stress response is a conjugation-deconjugation cycle that targets the transcription machinery. J Proteomics 118:39–48
Hendriks IA et al (2014) Uncovering global SUMOylation signaling networks in a site-specific manner. Nat Struct Mol Biol 21(10):927–936
Tammsalu T et al (2014) Proteome-wide identification of SUMO2 modification sites. Sci Signal 7(323):rs2
Hong Y et al (2001) Regulation of heat shock transcription factor 1 by stress-induced SUMO-1 modification. J Biol Chem 276(43):40263–40267
Goodson ML et al (2001) Sumo-1 modification regulates the DNA binding activity of heat shock transcription factor 2, a promyelocytic leukemia nuclear body associated transcription factor. J Biol Chem 276(21):18513–18518
Nathan D et al (2006) Histone sumoylation is a negative regulator in Saccharomyces cerevisiae and shows dynamic interplay with positive-acting histone modifications. Genes Dev 20(8):966–976
Kalocsay M, Hiller NJ, Jentsch S (2009) Chromosome-wide Rad51 spreading and SUMO-H2A.Z-dependent chromosome fixation in response to a persistent DNA double-strand break. Mol Cell 33(3):335–343
Shiio Y, Eisenman RN (2003) Histone sumoylation is associated with transcriptional repression. Proc Natl Acad Sci U S A 100(23):13225–13230
Miller MJ, Barrett-Wilt GA, Hua Z, Vierstra RD (2010) Proteomic analyses identify a diverse array of nuclear processes affected by small ubiquitin-like modifier conjugation in Arabidopsis. Proc Natl Acad Sci U S A 107(38):16512–16517
Issar N, Roux E, Mattei D, Scherf A (2008) Identification of a novel post-translational modification in Plasmodium falciparum: protein sumoylation in different cellular compartments. Cell Microbiol 10(10):1999–2011
Kouzarides T (2007) Chromatin modifications and their function. Cell 128(4):693–705
Dhall A et al (2014) Sumoylated human histone H4 prevents chromatin compaction by inhibiting long-range internucleosomal interactions. J Biol Chem 289(49):33827–33837
Dorigo B, Schalch T, Bystricky K, Richmond TJ (2003) Chromatin fiber folding: requirement for the histone H4 N-terminal tail. J Mol Biol 327(1):85–96
Kirsh O et al (2002) The SUMO E3 ligase RanBP2 promotes modification of the HDAC4 deacetylase. EMBO J 21(11):2682–2691
Bueno MT, Richard S (2013) SUMOylation negatively modulates target gene occupancy of the KDM5B, a histone lysine demethylase. Epigenetics 8(11):1162–1175
Hendriks IA, Treffers LW, Verlaan-de Vries M, Olsen JV, Vertegaal AC (2015) SUMO-2 orchestrates chromatin modifiers in response to DNA damage. Cell Rep. doi:10.1016/j.celrep.2015.02.033
Yang SH, Sharrocks AD (2004) SUMO promotes HDAC-mediated transcriptional repression. Mol Cell 13(4):611–617
Lindberg MJ, Popko-Scibor AE, Hansson ML, Wallberg AE (2010) SUMO modification regulates the transcriptional activity of MAML1. FASEB J 24(7):2396–2404
Murata T et al (2010) Transcriptional repression by sumoylation of Epstein-Barr virus BZLF1 protein correlates with association of histone deacetylase. J Biol Chem 285(31):23925–23935
Lyst MJ, Nan X, Stancheva I (2006) Regulation of MBD1-mediated transcriptional repression by SUMO and PIAS proteins. EMBO J 25(22):5317–5328
Ivanov AV et al (2007) PHD domain-mediated E3 ligase activity directs intramolecular sumoylation of an adjacent bromodomain required for gene silencing. Mol Cell 28(5):823–837
Nayak A, Viale-Bouroncle S, Morsczeck C, Muller S (2014) The SUMO-specific isopeptidase SENP3 regulates MLL1/MLL2 methyltransferase complexes and controls osteogenic differentiation. Mol Cell 55(1):47–58
Acknowledgements
The authors would like to thank Jen Gillies and Jason Berk for their helpful comments on the manuscript. They also acknowledge support from the US National Institutes of Health (R01 GM053756) to M.H. and an NIH Ruth L. Kirschstein National Research Service Award (NRSA) predoctoral fellowship (F31 AG046965) to N.R.W.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Wilson, N.R., Hochstrasser, M. (2016). The Regulation of Chromatin by Dynamic SUMO Modifications. In: Rodriguez, M. (eds) SUMO. Methods in Molecular Biology, vol 1475. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-6358-4_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-6358-4_2
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-6356-0
Online ISBN: 978-1-4939-6358-4
eBook Packages: Springer Protocols