Abstract
HIV-1/AIDS is often considered a priority disease in the development of genetic and cell based therapies because of the high burden imposed by current treatments, which require life-long adherence to antiretroviral drug regimens. Engineered nucleases have the capability to either disrupt a specific gene, or to promote precise gene edits or additions at the targeted gene. As one application for the gene disruption capabilities of the nucleases, HIV-1 infection provides an exceptional target in the CCR5 gene. This is the most commonly used entry co-receptor through which the virus enters into CD4+ T cells. Importantly, the loss of CCR5 is expected to be well-tolerated, since a relatively high percentage of individuals are naturally homozygous for a defective CCR5 allele. As a result, CCR5 disruption by zinc finger nuclease treatment of autologous T cells was the first-in-man use of engineered nucleases. Future applications to refine this therapy may include disrupting CCR5 in precursor hematopoietic stem cells, the additional disruption of the alternate HIV-1 co-receptor, CXCR4, in T cells, and the addition of other anti-HIV genes at a disrupted CCR5 locus to provide a combinatorial therapy. Finally, the gene disrupting actions of engineered nucleases could also be harnessed to inactivate the integrated HIV-1 genomes that persist in patients’ cells despite drug therapy, and which thereby prevent the complete eradication of the virus by drug treatments.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Homology directed repair (HDR)
- Hematopoietic stem cells (HSCs)
- Non-homologous end-joining (NHEJ)
- TAL effector nuclease (TALENs)
- Zinc finger nucleases (ZFNs)
Introduction
Human immunodeficiency virus (HIV-1) causes a serious, life-long infection, with high rates of mortality in untreated individuals. Although the current combinations of antiretroviral drugs used to treat the infection are highly effective at suppressing HIV-1 replication, they do not ultimately cure people. This means that infected individuals have a life-long requirement for antiretroviral drug therapy, with associated high economic costs and the risk of developing drug toxicities, viral resistance, or “treatment fatigue”. In fact, it is estimated that only 25 % of the HIV-infected population in the US successfully accesses therapy and achieves full viral suppression through antiretroviral drugs [1]. Therefore, alternate strategies to control and potentially cure HIV-1 infections are being considered, including those based on cell and genetic therapies.
HIV-1/AIDS has long been considered a candidate for gene therapy interventions, following early speculation that genetically modified cells could provide an ‘intracellular immunization’ to inhibit HIV-1 replication [2]. Some of the earliest gene therapy trials were for HIV-1, and typically used integrating retroviral vectors to allow for long-term expression of anti-HIV genes, such as the trans-dominant RevM10 protein and various RNA-based inhibitors (reviewed by Peterson et al. [3]). More recently, HIV-1 infection has proven itself uniquely suited to the first in-human use of engineered nucleases, based on CCR5 gene knockout by zinc finger nucleases (ZFNs) [4]. Beyond CCR5 disruption, future applications of engineered nucleases in anti-HIV therapies could exploit homologous recombination to insert anti-HIV genes at the disrupted CCR5 locus and thereby create a combinatorial gene therapy. Alternatively, the gene disrupting capabilities of the nucleases could be used to disable the integrated HIV-1 genomes present in infected cells, an attractive option for removing latently infected cells. The potential applications of engineered nucleases for HIV-1 therapy that will be discussed in this review are summarized in Fig. 1.
Potential uses of engineered nucleases as anti-HIV therapies. The main stages of the HIV-1 life-cycle are shown. HIV-1 enters a target cell by binding to both CD4 and a co-receptor protein such as CCR5 or CXCR4, the preference for which is determined by the HIV-1 surface glycoprotein. Following entry, the viral RNA genome is reverse transcribed into a DNA form that permanently integrates into the host cell’s genome. From here, the integrated provirus acts like a cellular gene, transcribing both mRNA to create HIV-1 proteins, and new viral RNA genomes, that are assembled together into particles that bud from the cell surface. (a) Engineered nucleases are used to disrupt the cellular co-receptor genes, CCR5 or CXCR4, in either CD4+ T cells (both genes), or their hematopoietic stem cell (HSC) precursors (CCR5 only) and thereby block HIV-1 entry. (b) Additional anti-HIV genes are inserted at the disrupted CCR5 locus, blocking other stages of the HIV-1 life-cycle. (c) Engineered nucleases are targeted to HIV-1 sequences, and thereby inactivate an integrated HIV-1 genome in an infected cell
Disruption of the CCR5 Co-Receptor by ZFNs
Engineered Nucleases and DNA Repair
Engineered nucleases such as zinc finger nucleases (ZFNs), TAL effector nucleases (TALENs), homing endonucleases and the CRIPSR/Cas9 system all function in basically the same way, by creating a double-stranded break (DSB) in the DNA sequence to which they are targeted, which is then acted on by cellular DNA repair pathways. While the homing endonucleases and Cas9 contain natural endonuclease activities, ZFNs and TALENs are based on a modular design that links engineered DNA binding domains to the non-specific cleavage domain from a homodimeric type IIS nuclease, such as the FokI restriction enzyme. Different cellular pathways can repair the DSBs so created, including the non-homologous end joining (NHEJ) pathway, where the frequent outcome is an insertion or deletion (indel) that can thereby lead to gene knockout (Fig. 2). Alternatively, DSBs can be more precisely repaired by recombination with a homologous sequence, such as a sister chromatid. Such homology directed repair (HDR) can also copy information from a homologous ‘donor sequence’, introduced into the cell at the same time as the engineered nuclease, and coding for any specific changes that are desired. The end result of this process can be a small genetic edit, for example to repair a point mutation in a defective gene, or the site-specific addition of a larger stretch of new genetic material at the site of the DSB. In this way, engineered nucleases can be used to direct three different outcomes: gene disruption, gene editing or gene addition (Fig. 2).
Alternate outcomes following the action of engineered nucleases. Engineered nucleases create a double-stranded break (DSB) in the targeted gene. If the non-homologous end joining (NHEJ) repair pathway is used, a frequent outcome is disruption of the gene. Alternatively, DSBs can be repaired by homologous recombination, and the introduction of a homologous donor sequence into the cell can hijack this pathway to introduce a desired genetic edit, or to promote the site-specific addition of new genetic material at the site of the DSB
The repair of a DSB is committed to one of the available cellular pathways at an early time point. If exposed DNA ends are protected by the Ku70/80 complex, the NHEJ pathway is used [5], while HDR is initiated if a resection event occurs that exposes tracts of single-stranded DNA [6]. The choice of HDR or NHEJ is also influenced by the phase of the cell cycle, with HDR only occurring during and shortly after DNA replication in S and G2 [6, 7], when the required factors are available in active (phosphorylated) forms [8], and sister chromatids are in closer proximity and more able to serve as homology templates [9]. In contrast, NHEJ predominates in G1, although it is active throughout the whole cell cycle.
In hematopoietic stem cells (HSC), an important therapeutic target cell for many gene therapy applications, NHEJ is far more common than HDR [10, 11], which biases the outcome of nuclease activities. In addition, the co-introduction of DNA donor sequences along with a nuclease, as is needed to promote HDR gene editing, can result in significant toxicity to these cells. Because of these factors, NHEJ-mediated gene disruption is the most easily achieved result for engineered nucleases in HSC. However, therapeutic applications of gene disruption are likely to be limited. In this regard, the application of engineered nucleases to HIV-1 disease has found a uniquely suitable target in the CCR5 gene (Fig. 2).
Rationale for CCR5 Disruption as an Anti-HIV Therapy
CCR5 is a chemokine receptor that also functions as the major entry co-receptor used by HIV-1, in concert with the primary receptor, CD4 [12] (Fig. 1). However, its functions are not essential in humans, since a relatively high frequency of the population (~1 %) is homozygous for the defective CCR5Δ32 allele. Such individuals are correspondingly almost completely resistant to HIV-1 [13, 14] while not exhibiting any significant phenotypic defects [13, 15]. These properties have encouraged the development of a class of anti-HIV drugs targeted to CCR5, such as Maraviroc [16], which further established CCR5 as a therapeutic target.
In addition, compelling human data exists for the ability of CCR5-negative cells to confer HIV-resistance, from the well-documented case of the so-called ‘Berlin patient ’. This individual was an HIV-positive leukemia patient who received an HSC transplant, as part of his leukemia treatment, from a donor who was homozygous for the CCR5Δ32 mutation. Following the treatment, the Berlin patient has been HIV-1 free for 8 years, and is considered the first documented case of an HIV-1 cure [17–19]. Although the treatment included aggressive ablative regimens to destroy the leukemia, which likely also significantly reduced the burden of HIV-infected cells, the essential role of the CCR5Δ32 genotype of the donor is illustrated by the fact that other HIV patients undergoing bone marrow transplantation without a CCR5Δ32 donor have not been similarly cured [20, 21]. These findings support the idea that blocking CCR5 expression in a patient’s own cells by genetic methods could also result in the control of HIV-1 replication.
Methods to block CCR5 have previously included the use of anti-CCR5 shRNAs or ribozymes, delivered using integrating retroviral or lentiviral vectors [22–27]. Such approaches can provide significant, albeit incomplete, inhibition, and will require the life-long expression of the anti-CCR5 moiety in the engineered cell. While the use of integrating vectors provides for this possibility, it is known that gene expression from such vectors can be silenced over time and that integrating viral vectors provide an unknown risk of insertional mutagenesis [28]. Because of these factors, there has been much interest in the use of engineered nucleases as an approach to disable the CCR5 gene. Since expression of the nucleases would only need to be a transient event, this approach can make use of potentially safer, non-permanent and non-integrating vector systems, yet still result in a permanent genetic change [29–35]. Both the mature CD4+ T cells that HIV-1 infects, as well as the precursor HSC that give rise to these cells, are considered suitable target cells for CCR5 engineering.
First to Clinic: CCR5 Knockout in T Cells Using ZFNs
CD4+ T cells are the primary target of HIV-1 infection, although other CD4+ cells such as macrophages are also infected by the virus. T cells are also an excellent clinical target cell for genetic manipulation, since considerable experience already exists from their use in anti-cancer applications, and the procedures to harvest, expand, genetically modify, store, and re-infuse these cells back into patients are well established [30, 36]. In addition, and contrary to initial expectations, genetically marked T cells have been shown to persist for at least 11 years in vivo, suggesting that any such modified T cells could confer a relatively long-lasting effect [37]. These factors made ZFN engineering of T cells for CCR5 disruption as an anti-HIV therapy an obvious first clinical application of engineered nucleases.
ZFNs against CCR5 were first described by Mani et al. [38], who constructed two different ZFN pairs targeted to the second and seventh transmembrane domains of CCR5 respectively (Fig. 3). Each ZFN pair consisted of monomers containing three zinc fingers, and was therefore capable of recognizing 9-bp target sequences on either side of the DSB site. Using these ZFNs, the authors were able to disrupt CCR5 plasmid DNA in an in vitro system. Since then, multiple studies have shown that ZFNs work well in a variety of human cells. Many of these studies have used a ZFN pair that creates a DSB centered at approximately nucleotide 160 in the open-reading frame, which maps to the first transmembrane domain of the CCR5 protein, and which contains four zinc fingers in each monomer [29–35]. Fortuitously, for a gene knockout approach, the most common indel resulting from this reagent is a 5 bp insertion that occurs in ~10–30 % of edited alleles and creates two in-frame stop codons shortly after the target site [31, 33, 35].
Target sites for engineered nucleases in CCR5. The approximate location of the target sites for a series of engineered nucleases are shown superimposed on a schematic of the CCR5 protein, and described more fully in the associated table. Also indicated is the location of the 32 bp deletion that produces a prematurely truncated and defective protein from the CCR5Δ32 allele. *Kim et al. [119] tested 315 ZFN pairs against 33 sites, and the 3 shown had the highest activity with lowest off-target effects. **Cho et al. [53] tested 10 CRISPRs against CCR5, and the one shown had the highest activity and lowest off-target effects. HE, homing endonuclease
The predicted ability of CCR5 disruption by ZFNs to impact HIV-1 replication has been demonstrated in an escalating series of studies in T cell lines [29, 31], primary T cell cultures [29–31, 39], and immune-deficient mice, engrafted with human T cells [29, 31, 39]. In the first such report, Perez et al. [31] showed that a CCR5 disruption rate of 2.4 % of alleles in the PM1 T cell line was increased to 73 % following infection of the culture with a CCR5-using (R5-tropic) strain of HIV-1. The authors further reported achieving CCR5 disruption rates between 28 and 33 % when the ZFNs were delivered to primary T cells using adenoviral vectors based on the Ad5/F35 variant [40]. When such modified T cells were engrafted into NOG mice and challenged with HIV-1, plasma viremia was reduced 7.2-fold compared to mice infused with unmodified T cells, and rates of CCR5 disruption at the end of the HIV-1 challenges increased by threefold (8.5–27.5 %). These pre-clinical studies demonstrated both the feasibility of using engineered nucleases to disrupt the CCR5 gene in primary human T cells, as well as the anti-HIV consequences of such engineering. The studies also formed the basis for a series of T cell based human clinical trials in HIV-infected individuals, using the same combination of the Ad5/F35 vector and the site 160 ZFN pair (Table 1).
The initial clinical trials of ZFN-mediated CCR5 disruption are based on a strategy of delivering CCR5 ZFNs to a patient’s own (autologous) T cells, which are then expanded ex vivo using CD3/CD28 stimulation. This protocol has been reported to allow the generation of up to 3 × 1010 T cells, with CCR5 disruption rates of 30–36 %, when measured at 10 days post transduction [30]. ZFNs were initially delivered as Ad5/35 vectors, but more recent trials are using mRNA electroporation. The first completed trial, NCT00842634, involved 12 patients who were each infused with 1010 T cells containing CCR5 modification rates between 10.9 and 27.7 % [4]. The procedure was well tolerated, with only one patient reporting an adverse reaction at time of infusion. CCR5-modified T cells were found to persist for several months, with a calculated half-life of 48 weeks. For 6 of the patients, a planned analytical treatment interruption (ATI) was initiated 4 weeks after infusion, involving cessation of antiretroviral therapy (ART) for 12 weeks, and this was completed by 4 of the 6 individuals. The rational for an ATI is that withdrawal of ART usually causes a rapid increase in HIV-1 viremia [41], and this may provide a selection pressure to increase the frequency of the CCR5-negative T cells. In addition, since this HIV-1 rebound follows a fairly characteristic pattern, an ATI provides an opportunity to evaluate whether the presence of CCR5-disrupted cells is influencing the ability of the patients to control HIV-1.
During the ATI, some indications of anti-HIV effects were observed. Although both CCR5-modified and unmodified CD4 T cells numbers declined in response to the rebound of HIV-1 viremia during the ATI, the rate of decline for the CCR5-modified T cells was significantly slower than the rate for the non-modified cells (−1.81 cells/mm3/day compared to −7.25 cells/mm3/day, p = 0.02), although this did not reach significance when mean values were considered (p = 0.08). In addition, in one of the patients undergoing ATI, HIV-1 RNA levels did not rebound until week 6 post ATI initiation, and then decreased to undetectable levels, even before therapy was re-started. Since this patient was heterozygous for the CCR5Δ32 allele, it is possible that this ‘half-way there’ genetic background facilitated the production of homozygous CCR5-negative cells by the ZFNs. This hypothesis is being further tested in clinical trial NCT01044654, in which 10 individuals who are CCR5Δ32 heterozygotes have been treated (Table 1). As a further refinement, the most recent T cell trials have also included treatment with cyclophosphamide (Cytoxan). This pre-conditioning treatment is expected to transiently reduce the patients’ T cell numbers prior to infusion of the ZFN-engineered cells, and thereby facilitate greater engraftment of the modified T cells.
CCR5 Disruption in HSC Using ZFNs
Engineered nucleases could also be used to disrupt the CCR5 gene in HSC, since these stem cells give rise to all lineages of the immune system, including CD4+ T cells. Although HSC are more challenging to work with than mature T cells, the longer life-span of the cells could allow for a one-shot treatment, while the fact that HSC give rise to both myeloid and lymphoid lineages means that non-T cell targets of HIV-1 such as macrophages would also be protected.
We previously reported on the ability of the site 160 ZFN pair to modify human HSC, isolated as CD34+ cells from umbilical cord blood [35]. By using plasmid DNA electroporation (Nucleofection) to introduce the ZFNs into the HSC, an average disruption rate of 17 % of the CCR5 alleles was achieved. These modified HSC were then used to engraft immune-deficient NSG mice, where they were found to differentiate comparably to unmodified HSC into various lineages of the human immune system, including CD4+ T cells. Following infection of the mice with R5-tropic HIV-1, protection of CD4+ T cells in the blood and lymphoid tissues was observed in the animals receiving ZFN-treated HSC compared to control HSC, which rapidly lost CD4+ T cells. Analysis of the blood and lymphoid tissues of the mice at 8–12 weeks post-infection also revealed a strong selection by HIV-1 for cells that were CCR5-negative. During the HIV-1 challenges, both unmodified and ZFN-engineered HSC cohorts of mice were equally infectable by HIV-1, but mice in the ZFN cohort were eventually able to suppress HIV-1, in both blood and tissues. This fits with the hypothesis that even a minority of CCR5-negative cells would be selected for during an active HIV-1 infection, and could eventually increase to a frequency where they were able to impact virus replication.
Although these observations support the use of ZFN-modified HSC as an alternative to engineering mature T cells, protocols will be needed that can deliver the nucleases to HSC while ensuring no or minimal impact on the ability of the cells to function as stem cells. In addition, although many pre-clinical studies rely on human HSC isolated from cord blood or fetal liver specimens, the clinical target cell for autologous HSC gene therapies will be the bone marrow resident HSC, which have distinct properties compared to other sources [42, 43]. These adult HSC are typically isolated by mobilization of the cells into the peripheral blood by treatment with cytokines (G-CSF), chemotherapeutic agents (cyclophosphamide), or small molecules such as the CXCR4 antagonist, AMD3100 [44]. We previously reported the modification of up to 25 % of CCR5 alleles in adult CD34+ HSC mobilized by G-CSF and treated with the same Ad5/F35 site 160 ZFN vectors as used in the current T cell trials (Table 1) [33]. However, adenoviral vector transduction of HSC proved to be more challenging than T cells, and transient low-dose treatment of the cells with PMA was necessary to achieve CCR5 disruption rates greater than 5 %. The associated toxicity cost when higher levels of gene disruption were achieved suggests that adenoviral vectors may not be optimal for this cell type.
In contrast, we have recently identified mRNA electroporation as a relatively non-toxic and efficient way to deliver ZFNs and TALENs to HSC, resulting in up to 50 % CCR5 disruption with minimal toxicity [45]. This method can also be used at large-scale, allowing a full patient dose of cells to be treated in one batch, and thereby increasing the clinical utility of this procedure.
CCR5 Disruption Using Other Engineered Nucleases
The recent progress to clinical trials of therapies based on ZFN modification of CCR5 is encouraging the development of other CCR5-specific reagents based on alternate engineered nucleases. The relative ease of design of TALENs compared to ZFNs is allowing the rapid evaluation of a range of CCR5-targeted TALENs, with variations in both the site of CCR5 that is targeted, as well as the basic design of the TALEN protein (e.g. length of the C-terminus). Miller et al. [46] first reported up to 15–20 % disruption of CCR5 in the K562 cell line using TALENs with either C28 or C63 backbone designs, respectively. For each backbone architecture, several left and right TALEN combinations were tested, allowing varying spacer lengths to be accommodated between the DNA sequences recognized by each monomer. All of the combinations evaluated were designed against sequences in the second extracellular domain of CCR5, close to the site of the natural Δ32 mutation (Fig. 3). Recently, we found that a C17 backbone TALEN also targeted close to the Δ32 site was able to achieve 60 % disruption in HSC [47]. Similarly, a C17 backbone TALEN targeting site 157, which overlaps the region recognized by the site 160 ZFNs, has also been evaluated by Mussolino et al., with the TALEN producing 17 % disruption of the CCR5 gene in 293 T cells [48]. Interestingly, when these authors looked at off-target disruption at the homologous CCR2 gene, they reported that such events were lower in cells transfected with the TALEN pair than the site 160 ZFN pair. In a follow up study, in which toxicity was more rigorously tested, the authors also noted lower overall cytotoxicity in the TALEN-treated cells [49]. Although both these TALENs and ZFNs targeted a similar region in CCR5, the non-identical nature of the target sequences recognized by the two reagents could have created such differences. In addition, while off-target and cytotoxicity analyses are a vital part of the evaluation of different candidate nucleases, especially for clinical applications, these analyses also need to be performed in the proposed clinical target cell, such as primary human T cells or HSC. Finally, it has been reported that CCR5 site 157 targeted TALENs can be introduced into cells by electroporation of mRNA, and thereby achieve disruption in both the PM1 cell line (up to 94 %) and primary T cells (between 17 and 46.8 %) [50].
It is possible that DNA breaks generated by different classes of nucleases will be qualitatively different, even when using the same FokI nuclease to introduce the break. For example, after analyzing a total of 1,456 mutant sequences at 122 target sites reported in 43 independent studies, Kim et al. found differences in the gene disruptions caused by ZFNs and TALENs. Specifically, they noted that while ZFNs cause an even distribution of insertions and deletions at the DSB site, TALENs almost exclusively cause deletions [51]. While the reason for this difference has not been determined, the authors speculated that the smaller spacer region between the two monomers in a ZFN pair may create a more defined cleavage site that is more prone to insertions, while the much larger spacers between the two regions bound by TALEN monomers could allow more heterogeneous cleavage sites, and result in more deletions. As noted earlier, the action of the widely used CCR5 site 160 ZFN pair frequently results in a 5 bp insertion that creates two in-frame stop codons, and which thereby terminates translation of the CCR5 protein
Homing endonucleases have also been adapted to target CCR5. These nucleases recognize stretches of 18–34 bp and cleave the target DNA in a manner similar to a restriction enzyme. Hundreds of homing endonucleases with different specificities have been reported, including I-Sce1, from baker’s yeast, and I-Cre1 from green algae chloroplasts. Although they can be very efficient at cleaving double stranded DNA, they do not use a specific code that is easily engineered to recognize a desired target site, as is the case for ZFNs or TALENs. This makes it more challenging to retarget them to specific sites such as CCR5. To date, one homing endonuclease has been reported that disrupts CCR5, based on I-CreI and targeting the third transmembrane domain of the protein (Fig. 3) [52]. The authors used combinatorial assembly of archived I-Cre1 derivatives to generate a final nuclease that matched the original I-Cre1 binding site at only 4 out of 24 bases. Interestingly, by co-expressing a DNA end-processing enzyme, the TREX2 exonuclease, the authors were also able to increase CCR5 disruption rates from 5 to 37 % in HSC, when both proteins were co-expressed from a lentiviral vector. This TREX2 co-expression also improved gene disruption rates for CCR5-targeted ZFNs and TALENs by almost threefold, suggesting a general approach to increasing gene disruption [52]. While the difficulty in engineering homing endonucleases is currently a bottleneck to their more widespread use, the small size of these proteins facilitates their delivery via viral vectors, which could make them attractive tools for certain gene therapy applications.
Finally, for the CRISPR/Cas9 system, since target site specificity is provided by a complementary RNA, these reagents are the simplest class of engineered nucleases to construct and evaluate, and lend themselves to high throughput analyses of different sites within a targeted gene. For example, Cho et al. [53] designed CRISPRs against 10 different sites in CCR5 and were able to disrupt up to 60 % of alleles in K562 cells using a CRISPR directed against the first extracellular domain of CCR5 (Fig. 3). Similarly, Cradick et al. described CRISPRs that could disrupt CCR5 at high levels (21–77 %) in 293T cells, targeting the N-terminal side of the second transmembrane domain and the second extracellular domain [54]. CRISPRs have also been shown to work in HSC. In a recent study, HSC were transfected with a Cas9 plasmid also expressing GFP. Cas9-expressing cells were then specifically FACS sorted based on GFP expression and electroporated with a CCR5 guide RNA plasmid. Subsequently, 25.8–30 % of colonies derived from these HSCs were found to have CCR5 disrupted at both alleles [55]. CRISPR/Cas9 components targeted to CCR5 have also been delivered via Ad5/F35 or lentiviral vectors: Li et al. were able to disrupt CCR5 at an average of 30.3 % of alleles in primary T cells using Ad5/F35 vectors [56], while Wang et al. achieved up to 43 % CCR5 negative cells when using lentiviral vector delivery to the CEMss-CCR5 T cell line [57].
CXCR4 is an Additional Target for Gene Knockout
Therapies based on engineered nucleases to disable the CCR5 co-receptor will always run into two potential limitations; (1) both copies of the gene need to be disabled to produce a CCR5-negative cell, and (2) certain strains of HIV-1 can use alternate co-receptors, such as CXCR4, to enter cells. While such X4-tropic viruses are not as common as R5-tropic strains, they emerge in ~50 % of HIV patients towards the later stages of the disease, and there is concern that therapies or drugs targeted towards CCR5 could speed this process or selection. This issue could be addressed by using CXCR4-targeted nucleases in addition to CCR5 reagents, although the important role that CXCR4 plays in HSC homing to the bone marrow stem cell niche means that CXCR4 disruption could only be feasible for T cells, and not HSC.
Targeting CXCR4 in T cells has been described by two groups. Wilen et al. showed that T cells modified by CXCR4 ZFNs were protected from X4-tropic HIV-1 infection in vitro, and that this provided partial protection in vivo in NSG mice transplanted with CXCR4 ZFN-modified T cells [58]. However, this protection was lost over time due to the emergence of R5-tropic strains of HIV-1. The authors also demonstrated that they could modify T cells obtained from homozygous CCR5Δ32 individuals, and that these cells were protected against both R5 and X4-tropic HIV-1. Yuan et al. further confirmed the finding that modification of CXCR4 with ZFNs protected cells in vitro and in vivo and additionally reported that this approach was more efficient at providing protection against X4-tropic HIV-1 than adenovirus vector delivery of shRNA against CXCR4 [59].
Taking this approach another step forward, Didigu et al. were able to co-transduce both a T cell line and primary T cells with two sets of ZFNs targeted against both CCR5 and CXCR4 [29]. These dual-disrupted T cells were rapidly enriched after infection with both R5 and X4-tropic viruses in vitro, confirming their protection. Consistent with the previous studies, this protection was also observed in vivo, when mice were transplanted with the T cells, previously infected with both R5 and X4-tropic viruses. Here, mice receiving the dual-ZFN treated T cells were found to have up to 200 times higher CD4 counts in the blood at day 55, compared to mice receiving unmodified cells, or those disrupted for CCR5 only [29]. This approach could therefore represent an improvement over disruption of CCR5 alone in T cells, and further suggests that CXCR4 disruption in T cells could be combined with CCR5 disruption in HSC. It should be noted, however, that while no toxic effects were observed in these model systems, the impact of disrupting CXCR4 in mature T cells is unknown, and further testing of this method will be required before use in humans.
Beyond Gene Knockout: Site-Specific Gene Addition and Combination Anti-HIV Therapies
As noted above, the ability of HIV-1 to adapt to use co-receptors other than CCR5 may limit the effectiveness of therapies relying solely on CCR5 disruption. Here, similar to the NIH guidelines for treatment with CCR5 inhibitor drugs [60], patients would first need to be screened to confirm that they do not already harbor viruses with X4 tropism. Beyond this limitation, CCR5 knockout is expected to only be effective in cells with homozygous disruptions, although a single allele knockout may mimic CCR5Δ32 heterozygotes, who do have a better prognosis than homozygous wild-type individuals [61–64]. Because of this limitation, the use of engineered nucleases to promote homologous recombination in order to insert additional anti-HIV genes into the CCR5 locus could provide a more effective strategy than CCR5 disruption alone (Fig. 1).
There are a number of anti-HIV molecules that are already being considered for more conventional HIV-1 gene therapies based on integrating retroviral and lentiviral vectors, and which could also be adapted for a gene knock-in approach. These include trans-dominant versions of HIV-1 proteins such as the RevM10 mutant [65, 66] and modified, HIV-resistant versions of cellular restriction factors such as TRIM5α and APOBEC3G. For example, although HIV-1 is naturally resistant to the human form of TRIM5α, it is inhibited by the Rhesus macaque ortholog [67], and engineered human-rhesus TRIM5α derivatives restore this anti-viral activity [68, 69]. Similarly, although the HIV-1 Vif protein normally degrades APOBEC3G, a cytosine deaminase that causes G→A mutations in the viral genome during reverse transcription, the D128K point mutant is resistant to Vif and thereby inhibits HIV-1 replication [70, 71]. Alternative candidates for insertion at the CCR5 locus include RNA based therapeutics such as ribozymes or small interfering RNAs (siRNA) directed against different HIV-1 targets such as Rev [72], Vif [73], Nef [74], Gag [75], Env [76, 77], or Tat [72, 73, 78], as well as the C46 peptide inhibitor that potently blocks HIV-1 fusion [79]. The feasibility of inserting three different anti-HIV genes (human-rhesus TRIM5α, APOBEC3G D128K, and Rev M10) at the CCR5 locus has been demonstrated in a cell line model [80].
For HIV-1 therapies and beyond, the use of site-specific gene addition in the primary human cells that would be used clinically will require significant increases in the efficiency of the process, which has proven to be challenging. In a pioneering study, Lombardo et al. [81] reported up to 50 % integration of a GFP cassette at the CCR5 site 160 in immortalized cell lines when using three non-integrating lentiviral vectors (IDLVs) to deliver each ZFN monomer and a homologous donor sequences. However, the difficulty of achieving such an outcome with human HSC was shown by the same vectors achieving only 0.11 % targeted GFP addition. Subsequently, the same group reported using adenovirus 5 vectors to deliver the CCR5 ZFNs, combined with IDLVs for the donor sequence, and were able to achieve up to 5 % targeted addition in primary human T cells at the site 160 locus [82].
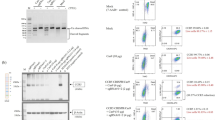
We have previously reported on the ability of site 160 ZFNs, delivered to human HSC as plasmid DNA, to achieve mean gene disruption rates of 17 % of CCR5 alleles [35]. When a CCR5 homology donor plasmid containing an internal PGK-GFP expression cassette was co-introduced into the cells with the ZFNs, although we observed an increase in DNA-mediated toxicity, the viable human HSC in the population were able to engraft NSG mice, and give rise to mature human cells stably expressing GFP at frequencies up to 5 % in multiple lineages (Fig. 4a). When humanized mice are challenged with an R5-tropic strain of HV-1, the HIV-mediated destruction of CD4 T cells typically causes selection of phenotypically CCR5-negative cells, created by the action of the nucleases [35]. In this experimental situation where GFP was inserted at the CCR5 locus, HIV-1 infection also co-selected for GFP-expressing cells in the CD4 cell fraction only (Fig. 4a, b). This observation is in keeping with the GFP cassette being specifically integrated at the CCR5 locus, which was also confirmed by PCR analysis (Fig. 4c).
Targeted gene addition at the CCR5 locus. Human cord blood HSC were electroporated with plasmid DNAs expressing CCR5 site 160 ZFNs and a donor sequence containing a PGK-GFP cassette flanked by CCR5 homologous sequences. The cells were engrafted into neonatal NSG mice, as described previously [35]. At 12 weeks of age, the mice were infected with R5-tropic HIV-1. (a) At the indicated time points, blood was analyzed by FACS for GFP expression in total human leukocytes (CD45 cells), and in the CD4 and CD8 subsets. (b) Following HIV-1 infection, the frequency of GFP-expressing cells increased in the CD4 fraction, which represent the target cells of HIV-1 infection, as well as the total CD45 group, but not in the CD8 fraction. (c) The site-specific addition of the GFP cassette at the CCR5 locus was confirmed by a specific in-out PCR analysis of clones derived from the input HSC, where one of the PCR primers is specific for the GFP expression cassette, and one is specific for CCR5, but beyond the region contained in the donor sequence
Despite these promising developments, and the expected ability of HIV-1 itself to act as a selection agent to increase the frequency of cells with CCR5-specific gene addition, the currently achievable levels of targeted addition in HSC cannot match the efficiency of more standard integrating viral vectors for the delivery of genes, and higher rates of targeted integration, especially into primary HSC, may be required. Recent promising progress has been made by introducing homologous donors using non-integrating lentiviral [83, 84] and AAV vectors [85] and strategies that can promote HDR over NHEJ in human cells are also being explored [86, 87].
Disrupting CCR5 in iPSC
HSC are challenging to culture and expand in vitro without losing their ability to differentiate, which limits the potential to pre-select genome modified cells prior to infusion into a patient. In the future, it may be possible to derive fully functional HSC from patient-specific induced pluripotent stem cells (iPSC), which would thereby also allow for the generation of a more homogenous population of modified cells. Towards this goal, several groups have recently reported modifying iPSC: Ye et al. used CRISPRs and TALENs to generate iPSC that contained the exact CCR5Δ32 mutation while maintaining pluripotency [88], while Yao et al. disrupted CCR5 in human embryonic stem cells and iPSC and were able to differentiate them into CD34+ cells and more differentiated hematopoietic lineages [89].
Disrupting HIV-1 Genomes with Engineered Nucleases
Progress to Developing HIV-Specific Nucleases
An additional application of engineered nucleases to combat HIV-1 would be to engineer the reagents to target the HIV-1 genome itself (Fig. 1). Although antiretroviral drugs are highly effective and capable of suppressing HIV-1 to undetectable levels, they do not completely eliminate the virus, which then usually rebounds if therapy is stopped. The source of this rebounding virus are the so-called latent HIV reservoirs—cells in the body where the virus lies dormant, unperturbed by antiretroviral drugs, but from which the virus can be activated at a later date. Among the most prominent reservoirs are memory CD4 T cells [90–92]. Methods to remove or disable the virus in these cells could allow drug-free suppression of HIV-1, or even a cure.
There have been several reports in which different types of engineered nucleases have been developed that recognize the HIV-1 genome [93–96]. In most of these studies, the targeted sequence has been the HIV-1 long terminal repeat (LTR), which is present at both ends of the integrated DNA version of the virus. The LTRs play multiple roles, with the 5′ sequence driving transcription of the viral genome and the 3′ sequence regulating transcription termination and polyadenylation. Intact LTRs are also necessary for correct RNA processing to allow successful reverse transcription and integration during the viral life-cycle. In addition, the presence of two copies of the LTR in the HIV-1 genome provides anuclease with double the target sequences, as well as the possibility of a double cut and re-ligation event, leading to the permanent excision of the intervening HIV-1 genome.
One of the first reports describing disruption of an integrated HIV-1 genome through engineering recognition of specific HIV-1 sequences used a modified cre-recombinase [93]. This enzyme usually recognizes 34 bp loxP sites, but was evolved to recognize a similar sequence that is present in the LTRs of the clade A HIV-1 strain, TZB0003 [97]. The evolved recombinase, named Tre recombinase, was able to disrupt integrated HIV-1 genomes in HeLa cells, and reduce the production of new HIV-1 particles [93]. The Tre recombinase was subsequently evaluated in an in vivo model [95], where primary T cells or HSC were transduced with lentiviral vectors expressing the recombinase under control of an HIV-inducible promoter. This design used a Tre-resistant HIV-1 LTR promoter containing tandem copies of the HIV-1 Tar element, which responds to the viral transactivator, Tat, produced upon HIV-1 infection. Consequently, the recombinase would only be expressed in cells infected with HIV-1, so reducing the potential for toxicity. In this proof of concept experiment, cells transduced with the vectors at between 30 and 60 % were engrafted into Rag2-/-γc-/-mice, which were then challenged with a replication-competent R5-tropic HIV-1 reporter virus, modified to contain the Tre recognition sites in its LTRs. The animals receiving the Tre recombinase-containing cells had increasing or stable levels of human CD45 and CD4 cells, and virus levels that decreased approximately 1-log between weeks 2 and 12 post infection. This was in contrast to mice receiving non-transduced cells, which showed decreasing levels of human CD45 and CD4 cells and increasing or stable HIV-1 levels over the same time period. While this is an intriguing approach, the broader application of the technology will require the development of recombinases that recognize sequences that are more divergent from the natural loxP sequence and, preferably, are highly conserved across multiple strains of HIV-1. Such engineering challenges may be facilitated by the availability of new design tools [98]. In this regard, another Cre recombinase derivative (uTre) has recently been described that recognizes an HIV LTR sequence that is conserved in 94 % of the major HIV subtypes A, B, and C, which would make it much more therapeutically relevant [99].
For the classes of engineered nucleases with less restrictions on target site selection (ZFNs, TALENs and CRISPR/Cas9), several different regions of the HIV-1 genome could provide target sites in addition to the LTRs. Figure 5 displays an entropy analysis of sequence variation in the HIV-1 genome, highlighting regions of conservation that could provide a source of such target sites. ZFNs have already been described targeting the LTRs, using a site that is highly conserved in viral isolates from patient samples [94]. These reagents were reported to disrupt up to 60 % of GFP-expressing HIV-1 genomes present in a Jurkat T cell line following DNA electroporation, and to reduce viral production from infected primary human PBLs (29 % reduction) or CD4+ T cells (31 % reduction). Similarly, TALENs have also been used to target HIV DNA. Ebina et al. were able to significantly disrupt HIV sequences in a Jurkat cell line model of an integrated latent HIV genome, where LTR-driven GFP expression occurs after stimulation with TNFα. By electroporating in mRNA for a TALEN pair targeting the LTR, they saw reduction in the levels of GFP-positive cells following stimulation from 63 % down to 4.3 %, and reported complete removal of the HIV genome in 53 % of the cells [100].
Identification of low entropy islands in the HIV-1 genome. 102 LTR and 442 coding region sequences from the Los Alamos HIV-1 sequence database were queried using the Entropy-one tool (www.hiv.lanl.gov). Entropy measures the amount of sequence variation at each position, with lower entropy scores indicating higher conservation. The average entropy score across sequential 20 bp segments was calculated and plotted, to highlight islands (red lines) of consistent low entropy (at or below 0.1)
Finally, CRISPRs are also being used to target integrated HIV-1 DNA. Here, the potential for these reagents to be multiplexed [101] and thereby target more than one HIV-1 sequence could prove to be a distinct advantage against a virus known for its ability to rapidly evolve resistance to other therapies. Ebina et al. designed two CRISPR gRNAs against the TAR and NFκB regions of the LTR, which were tested for their ability to block expression from an integrated LTR-GFP reporter sequence that also contained the Tat gene (JLAT cells) [96]. The utility of the reagents to disrupt LTR function was demonstrated following three rounds of electroporation with the CRISPR/Cas9 reagents, where GFP expression was reduced in activated cells by 25–32 % [96]. Similarly, Zhu et al. tested 10 different gRNAs against the HIV LTR and Pol regions in JLAT cells and were able to reduce the levels of GFP+ cells by more than 24-fold when used in combination [102]. Liao et al. also found that introducing multiple CRISPR gRNAs was better able to disrupt integrated HIV sequences when compared to single gRNAs, and they further demonstrated that cells expressing anti-HIV CRISPRs were protected from HIV infection [103]. Finally, Hu et al. reported an effect in CHME-5 cells, a microglial cell line of HIV latency, where GFP expression after activation was reduced from 76 to 17.1 % or 3.9 % of cells for two different LTR-directed gRNAs [104].
Challenges for the Use of Anti-HIV Nucleases
While the studies described above represent important first steps toward using engineered nucleases to disrupt HIV-1, much work will be needed to deliver these reagents to the cells in patients that harbor latent HIV-1 genomes, including those cells where latency may involve chromatin condensation. In addition, even when highly conserved HIV-1 sequences are targeted (Fig. 5), the potential for HIV-1 to mutate and evolve resistance against other therapies is well established, and it should be assumed that this will also be the case for HIV-specific engineered nucleases. In this regard, targeting multiple sequences simultaneously may be advantageous, including all known variants known to be tolerated at a specific site, although this would greatly increase the complexity of the therapy. In addition, in common with all therapeutic applications using nucleases, the potential for off-target effects will need to be characterized. Finally, unless a lie-in-wait protection approach is to be used, as described above for the Tat-inducible Tre-recombinase system, methods to deliver the reagents to HIV-infected cells in vivo will need to be developed. This requirement will be especially challenging for latently infected cells, since the lack of expression of HIV-1 genes means that there are no obvious signs to indicate that the cells harbor an HIV-1 genome. Thus, bulk delivery to all T cells, or select delivery through the use of surrogate markers of reservoir cells [90], may be needed.
Considerations for the Clinical Use of Engineered Nucleases
Strategies to Deliver Engineered Nucleases to Human Cells
The delivery of engineered nucleases to cells presents certain challenges, including the requirement for the co-delivery of more than one component for the ZFN, TALEN and CRISPR/Cas9 nucleases, and the additional inclusion of a donor sequence if HDR-mediated engineering is desired. Beyond this, the extensive regions of repeat sequences present in the DNA binding domains of ZFNs and TALENs can lead to instability. However, since the permanent genetic changes that result from the action of engineered nucleases only require their transient expression in target cells, a wide variety of delivery systems could potentially be used.
Reflecting their earlier development and adoption for clinical applications, ZFNs are the class of engineered nucleases that have been evaluated in the widest array of delivery methods. These include packaging in standard viral vectors such as adenovirus, adeno-associated virus (AAV) and IDLVs [31, 33, 81, 82, 105–107] as well as less frequently used systems such as baculovirus vectors [32]. In a simpler delivery approach, nucleases have also been introduced into cells following electroporation of both DNA and mRNA [32, 35, 45, 47, 81, 82]. Interestingly, ZFN proteins are also capable of direct uptake by cells, with one study reporting CCR5 disruption rates up to 27 % in cell lines, and 8 % in primary human CD4 T cells, using this approach [34]. The authors also reported lower rates of off-target disruption at 9 sites with homology to CCR5 when compared to the rates occurring following transfection of ZFN DNA into 293T cells although, as noted, this may reflect lower concentrations of the ZFNs when delivered by this route. In a similar approach, Chen et al. developed a system whereby ZFN proteins are attached to the transferrin molecule through a cleavable disulfide bond [108]. Here, the ZFN proteins are taken up into cells by transferrin receptor-mediated endocytosis, and the system appears to function in a variety of cell types, reflecting the broad distribution of this receptor. Such protein-based delivery systems may be simpler to use than viral based systems and, by reducing the time of exposure of the cells to ZFN proteins, may indeed reduce off-target effects, which are a function of both the concentration of the nucleases, and the time of exposure.
Although TALENs and ZFNs share many common design features, TALEN delivery is likely to be more complicated than ZFNs. First, TALEN constructs with the commonly used C63 backbone are roughly 1 kb larger (per monomer) than ZFNs, which complicates their use in size constrained vectors such as AAV. In addition, the 1 base pair recognition code used by the TALE repeat component is in contrast to the 3 bp recognition sequence of each zinc finger module, so that TALENs directed to the same target sequence contain three times as many repeating units as ZFNs. Such designs can cause instability in certain vector types, especially lentiviral vectors [107], where the high level of homology between the TALEN subunits can lead to rearrangements and deletions as a result of strand switching between the subunits of the two packaged vector RNAs during reverse transcription. A possible solution to this problem will be to make numerous silent mutations in each of the repeat units of the TALE to decrease homology. Alternatively, it was found in the same study that Ad5 vectors worked well for delivering TALENs, producing 47–55 % gene disruption of the AAVS1 locus in HeLa cells, immortalized myoblasts and bone marrow-derived mesenchymal stem cells [107]. Recently another option was described in which the lentiviral vector delivering the TALEN was mutated to be unable to perform reverse transcriptase. The resulting RNA genome was able to express an encoded TALEN following addition of an IRES and poly A sequence, to mimic a mRNA [109].
For all classes of engineered nucleases, current technologies are limited almost exclusively to methods of ex vivo delivery , but future broader utilization of the reagents will benefit from in vivo methods of delivery, as are being developed for siRNAs [110], and may be possible for certain viral vectors. For example, different subtypes of AAV display different in vivo tropisms, and AAV vectors have been shown to be capable of co-delivering both ZFN monomers and a donor sequence to mouse hepatocytes following intraperitoneal injection of hepatotropic AAV vectors [105]. In addition, recent advances in retargeting lentiviral vectors, based on including scFvs or ligands to cell surface proteins, could be exploited to allow the delivery of engineered nucleases to specific cell types in the future [111–117].
Off-Target Effects and Toxicity
A major concern for any clinical application of engineered nucleases is the potential for adverse events, including cellular toxicity [118], or nuclease modification at off-target sites [31, 33, 119–121]). Toxicity may result from expression of the nucleases themselves, be a consequence of the cellular response to DNA damage, or result from a combination of factors that stress the cells, including the delivery method used. Off-target gene modifications can occur at sites predicted by bioinformatics to be highly homologous to the desired target sequence, as well as at sites that are revealed by in vitro screens [122], or by DSB site capture in cells following NHEJ-mediated integration of IDLVs [123]. Since such off-target events and toxicities may be cell-type specific, cell line studies may not predict the outcome for primary human cells. Thus pre-clinical toxicology studies should include both the evaluation of activity at predicted off-target sites, as well as more general analyses to evaluate the tumorigenic potential of patient-sized doses of ZFN-treated T cells or HSC, using both in vitro cellular assays and following engraftment into immune-deficient mice [30, 42].
Recently, several new methods to detect off-target events have been described. For example, high-throughput genome-wide translocation sequencing (HTGTS) was used to identify translocation junctions in cells treated with I-sce meganuclease [124] and this method has been adapted and enhanced by Frock et al. to include linear-amplification-mediated PCR (LAM-PCR) to identify off-target sites created by CRISPRs and TALENs [125]. This study confirmed previous findings that CRISPR-mediated off-target activity can be reduced by using CRISPR nickases [53], and also found that TALEN-mediated off-target effects were mostly due to homodimers [125].
Other genome-wide off-target detection methods include GUIDE-seq (genome-wide, unbiased identification of DSBs enabled by sequencing), which is based on detection of double-stranded oligodeoxynucleotide tags inserted into the DSBs [126], and the BLESS assay (breaks labeling, enrichments on streptavidin and next-generation sequencing), which labels DSBs with biotinylated oligonucleotides [127, 128]. Yet another method is digenome-seq (digested genome sequencing), which works by nuclease treatment of genomic DNA in vitro, followed by whole genome sequencing of the resulting fragments [129]. The value of these assays is that they don’t rely on bioinformatics predictions and so can potentially detect cryptic off-target sites in a more unbiased way. However, the assays are still imperfect since the HTGTS, GUIDE-seq and digenome-seq all produced different sets of off-target sites for a VEGF-A CRISPR target site (Reviewed in [120]).
For the commonly used site 160 CCR5 ZFNs, off-target disruption is most commonly observed at the highly homologous CCR2 gene, and can reach 9–10 % of the rates at the CCR5 locus [31, 33, 49, 119, 122, 123], and increasing as the expression of ZFNs is increased. In this way, it may be useful to consider nuclease activity as having a practical plateau, with a maximum amount of on-target disruption and acceptable amount of off-target activity. Meanwhile, methods to reduce off-target activity of nucleases are also being developed. For example, both ZFNs and TALENs are made more specific through the use of engineered obligate heterodimers of the Fok1 endonuclease, which limits the possible pair combinations of the individual component monomers to the desired heterodimer [130]. In a related approach, CRISPR/Cas9 specificity can be improved by requiring two guide RNA binding events and using a catalytically inactive Cas9 protein fused to Fok1 moieties [131]. Finally, altering the nature of the DNA-binding RVDs in TALENs can also result in reduced off-target effects by increasing on-target specificity [47].
Summary
The life-cycle of HIV-1 provides several opportunities for interventions by therapies based on engineered nucleases. The requirement of the virus for a cellular co-receptor, CCR5, that is non-essential to its human host, provides an obvious application for the gene disrupting capabilities of nucleases, and the use of ZFNs in this regard is currently the most clinically advanced application of this new class of genome modifying tools. Beyond that, gene disruption could be combined with HDR to insert additional anti-HIV genes at the CCR5 locus. In addition, the integrated HIV-1 genomes that persist in patients’ cells despite antiretroviral therapy can also be considered as a genetic target for disruption, although the challenges of delivering nucleases to the cells that harbor such latent genomes will be formidable. Finally, as in all gene therapy approaches that create HIV-resistant cells, it is anticipated that HIV-1 could be harnessed to assist in its own demise, by enabling selection for the engineered, HIV-resistant cells.
References
http://aids.gov/federal-resources/policies/care-continuum, 2013.
Baltimore D. Gene therapy. Intracellular immunization. Nature. 1988;335(6189):395–6.
Peterson CW, et al. Combinatorial anti-HIV gene therapy: using a multipronged approach to reach beyond HAART. Gene Ther. 2013;20(7):695–702.
Tebas P, Stein D, Tang WW, Frank I, Wang SQ, Lee G, et al. Gene editing of CCR5 in autologous CD4 T cells of persons infected with HIV. N Engl J Med. 2014;370:10.
Sun J, et al. Human Ku70/80 protein blocks exonuclease 1-mediated DNA resection in the presence of human Mre11 or Mre11/Rad50 protein complex. J Biol Chem. 2012;287(7):4936–45.
Symington LS, Gautier J. Double-strand break end resection and repair pathway choice. Annu Rev Genet. 2011;45:247–71.
Chapman JR, Taylor MR, Boulton SJ. Playing the end game: DNA double-strand break repair pathway choice. Mol Cell. 2012;47(4):497–510.
Escribano-Diaz C, et al. A cell cycle-dependent regulatory circuit composed of 53BP1-RIF1 and BRCA1-CtIP controls DNA repair pathway choice. Mol Cell. 2013;49(5):872–83.
Heyer WD, Ehmsen KT, Liu J. Regulation of homologous recombination in eukaryotes. Annu Rev Genet. 2010;44:113–39.
Mohrin M, et al. Hematopoietic stem cell quiescence promotes error-prone DNA repair and mutagenesis. Cell Stem Cell. 2010;7(2):174–85.
Rossi DJ, et al. Hematopoietic stem cell quiescence attenuates DNA damage response and permits DNA damage accumulation during aging. Cell Cycle. 2007;6(19):2371–6.
Deng H, et al. Identification of a major co-receptor for primary isolates of HIV-1. Nature. 1996;381(6584):661–6.
Samson M, et al. Resistance to HIV-1 infection in Caucasian individuals bearing mutant alleles of the CCR-5 chemokine receptor gene. Nature. 1996;382(6593):722–5.
Novembre J, Galvani AP, Slatkin M. The geographic spread of the CCR5 Delta32 HIV-resistance allele. PLoS Biol. 2005;3(11), e339.
Dean M, et al. Genetic restriction of HIV-1 infection and progression to AIDS by a deletion allele of the CKR5 structural gene. Hemophilia Growth and Development Study, Multicenter AIDS Cohort Study, Multicenter Hemophilia Cohort Study, San Francisco City Cohort, ALIVE Study. Science. 1996;273(5283):1856–62.
Wood A, Armour D. The discovery of the CCR5 receptor antagonist, UK-427,857, a new agent for the treatment of HIV infection and AIDS. Prog Med Chem. 2005;43: 239–71.
Allers K, et al. Evidence for the cure of HIV infection by CCR5Delta32/Delta32 stem cell transplantation. Blood. 2011;117(10):2791–9.
Yukl SA, et al. Challenges in detecting HIV persistence during potentially curative interventions: a study of the Berlin patient. PLoS Pathog. 2013;9(5), e1003347.
Hutter G, et al. Long-term control of HIV by CCR5 Delta32/Delta32 stem-cell transplantation. N Engl J Med. 2009;360(7):692–8.
Hutter G, Zaia JA. Allogeneic haematopoietic stem cell transplantation in patients with human immunodeficiency virus: the experiences of more than 25 years. Clin Exp Immunol. 2011;163(3):284–95.
Hayden EC. Hopes of HIV cure in ‘Boston patients’ dashed. Nature News 2013.
Ringpis GE, et al. Engineering HIV-1-resistant T-cells from short-hairpin RNA-expressing hematopoietic stem/progenitor cells in humanized BLT mice. PLoS One. 2012;7(12), e53492.
Lee MT, et al. Inhibition of human immunodeficiency virus type 1 replication in primary macrophages by using Tat- or CCR5-specific small interfering RNAs expressed from a lentivirus vector. J Virol. 2003;77(22):11964–72.
ter Brake O, et al. Lentiviral vector design for multiple shRNA expression and durable HIV-1 inhibition. Mol Ther. 2008;16(3):557–64.
Qin XF, et al. Inhibiting HIV-1 infection in human T cells by lentiviral-mediated delivery of small interfering RNA against CCR5. Proc Natl Acad Sci U S A. 2003;100(1):183–8.
An DS, et al. Optimization and functional effects of stable short hairpin RNA expression in primary human lymphocytes via lentiviral vectors. Mol Ther. 2006;14(4):494–504.
Li MJ, et al. Inhibition of HIV-1 infection by lentiviral vectors expressing Pol III-promoted anti-HIV RNAs. Mol Ther. 2003;8(2):196–206.
Wu C, Dunbar CE. Stem cell gene therapy: the risks of insertional mutagenesis and approaches to minimize genotoxicity. Front Med. 2011;5(4):356–71.
Didigu CA, et al. Simultaneous zinc-finger nuclease editing of the HIV coreceptors ccr5 and cxcr4 protects CD4+ T cells from HIV-1 infection. Blood. 2014;123(1):61–9.
Maier DA, et al. Efficient clinical scale gene modification via zinc finger nuclease-targeted disruption of the HIV co-receptor CCR5. Hum Gene Ther. 2013;24(3):245–58.
Perez EE, et al. Establishment of HIV-1 resistance in CD4+ T cells by genome editing using zinc-finger nucleases. Nat Biotechnol. 2008;26(7):808–16.
Lei Y, et al. Gene editing of human embryonic stem cells via an engineered baculoviral vector carrying zinc-finger nucleases. Mol Ther. 2011;19(5):942–50.
Li L, et al. Genomic editing of the HIV-1 coreceptor CCR5 in adult hematopoietic stem and progenitor cells using zinc finger nucleases. Mol Ther. 2013;21(6):1259–69.
Gaj T, et al. Targeted gene knockout by direct delivery of zinc-finger nuclease proteins. Nat Methods. 2012;9(8):805–7.
Holt N, et al. Human hematopoietic stem/progenitor cells modified by zinc-finger nucleases targeted to CCR5 control HIV-1 in vivo. Nat Biotechnol. 2010;28(8):839–47.
Kalos M, June CH. Adoptive T cell transfer for cancer immunotherapy in the era of synthetic biology. Immunity. 2013;39(1):49–60.
Scholler J, et al. Decade-long safety and function of retroviral-modified chimeric antigen receptor T cells. Sci Transl Med. 2012;4(132):132ra53.
Mani M, et al. Design, engineering, and characterization of zinc finger nucleases. Biochem Biophys Res Commun. 2005;335(2):447–57.
Yi G, et al. CCR5 gene editing of resting CD4(+) T cells by transient ZFN expression from HIV envelope pseudotyped nonintegrating lentivirus confers HIV-1 resistance in humanized mice. Mol Ther Nucleic Acids. 2014;3, e198.
Shayakhmetov DM, et al. Efficient gene transfer into human CD34(+) cells by a retargeted adenovirus vector. J Virol. 2000;74(6):2567–83.
Chun TW, et al. Relationship between pre-existing viral reservoirs and the re-emergence of plasma viremia after discontinuation of highly active anti-retroviral therapy. Nat Med. 2000;6(7):757–61.
Hofer U, et al. Pre-clinical modeling of CCR5 knockout in human hematopoietic stem cells by zinc finger nucleases using humanized mice. J Infect Dis. 2013;208 Suppl 2:S160–4.
Lepus CM, et al. Comparison of human fetal liver, umbilical cord blood, and adult blood hematopoietic stem cell engraftment in NOD-scid/gammac−/−, Balb/c-Rag1−/−gammac−/−, and C.B-17-scid/bg immunodeficient mice. Hum Immunol. 2009;70(10):790–802.
Lapid K, et al. Egress and mobilization of hematopoietic stem and progenitor cells: a dynamic multi-facet process. Cambridge: Harvard Stem Cell Institute; 2008. StemBook [Internet].
Cannon PM, et al. Electroporation of ZFN mRNA enables efficient CCR5 gene disruption in mobilized blood hematopoietic stem cells at clinical scale. Mol Ther. 2013;21:S71–2.
Miller JC, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2011;29(2):143–8.
Llewellyn N, et al. Next generation TALENs mediate efficient disruption of the CCR5 gene in human HSCs. Mol Ther. 2013;21:S72.
Mussolino C, et al. A novel TALE nuclease scaffold enables high genome editing activity in combination with low toxicity. Nucleic Acids Res. 2011;39(21):9283–93.
Mussolino C, et al. TALENs facilitate targeted genome editing in human cells with high specificity and low cytotoxicity. Nucleic Acids Res. 2014;42(10):6762–73.
Mock U, et al. mRNA transfection of a novel TAL effector nuclease (TALEN) facilitates efficient knockout of HIV co-receptor CCR5. Nucleic Acids Res. 2015;43(11):5560–71.
Kim Y, Kweon J, Kim JS. TALENs and ZFNs are associated with different mutation signatures. Nat Methods. 2013;10(3):185.
Certo MT, et al. Coupling endonucleases with DNA end-processing enzymes to drive gene disruption. Nat Methods. 2012;9(10):973–5.
Cho SW, et al. Analysis of off-target effects of CRISPR/Cas-derived RNA-guided endonucleases and nickases. Genome Res. 2014;24(1):132–41.
Cradick TJ, et al. CRISPR/Cas9 systems targeting beta-globin and CCR5 genes have substantial off-target activity. Nucleic Acids Res. 2013;41(20):9584–92.
Mandal PK, et al. Efficient ablation of genes in human hematopoietic stem and effector cells using CRISPR/Cas9. Cell Stem Cell. 2014;15(5):643–52.
Li C, et al. Inhibition of HIV-1 infection of primary CD4+ T cells by gene editing of CCR5 using adenovirus-delivered CRISPR/Cas9. J Gen Virol. 2015;96(8):2381–93.
Wang W, et al. CCR5 gene disruption via lentiviral vectors expressing Cas9 and single guided RNA renders cells resistant to HIV-1 infection. PLoS One. 2014;9(12), e115987.
Wilen CB, et al. Engineering HIV-resistant human CD4+ T cells with CXCR4-specific zinc-finger nucleases. PLoS Pathog. 2011;7(4), e1002020.
Yuan J, et al. Zinc-finger nuclease editing of human cxcr4 promotes HIV-1 CD4(+) T cell resistance and enrichment. Mol Ther. 2012;20(4):849–59.
http://aidsinfo.nih.gov/guidelines/html/1/adult-and-adolescent-arv-guidelines/8/co-receptor-tropism-assays. Guidelines for the use of antiretroviral agents in HIV-1-infected adults and adolescents. 2013.
Meyer L, et al. Early protective effect of CCR-5 delta 32 heterozygosity on HIV-1 disease progression: relationship with viral load. The SEROCO Study Group. AIDS. 1997;11(11):F73–8.
de Roda Husman AM, et al. Association between CCR5 genotype and the clinical course of HIV-1 infection. Ann Intern Med. 1997;127(10):882–90.
Meyer L, et al. CCR5 delta32 deletion and reduced risk of toxoplasmosis in persons infected with human immunodeficiency virus type 1. The SEROCO-HEMOCO-SEROGEST Study Groups. J Infect Dis. 1999;180(3):920–4.
Ioannidis JP, et al. Effects of CCR5-Delta, 32, CCR2–64I, and SDF-1 3′A alleles on HIV-1 disease progression: an international meta-analysis of individual-patient data. Ann Intern Med. 2001;135(9):782–95.
Bevec D, et al. Inhibition of human immunodeficiency virus type 1 replication in human T cells by retroviral-mediated gene transfer of a dominant-negative Rev trans-activator. Proc Natl Acad Sci U S A. 1992;89(20):9870–4.
Bonyhadi ML, et al. RevM10-expressing T cells derived in vivo from transduced human hematopoietic stem-progenitor cells inhibit human immunodeficiency virus replication. J Virol. 1997;71(6):4707–16.
Stremlau M, et al. The cytoplasmic body component TRIM5alpha restricts HIV-1 infection in Old World monkeys. Nature. 2004;427(6977):848–53.
Sawyer SL, et al. Positive selection of primate TRIM5alpha identifies a critical species-specific retroviral restriction domain. Proc Natl Acad Sci U S A. 2005;102(8):2832–7.
Anderson J, Akkina R. Human immunodeficiency virus type 1 restriction by human-rhesus chimeric tripartite motif 5alpha (TRIM 5alpha) in CD34(+) cell-derived macrophages in vitro and in T cells in vivo in severe combined immunodeficient (SCID-hu) mice transplanted with human fetal tissue. Hum Gene Ther. 2008;19(3):217–28.
Schrofelbauer B, Chen D, Landau NR. A single amino acid of APOBEC3G controls its species-specific interaction with virion infectivity factor (Vif). Proc Natl Acad Sci U S A. 2004;101(11):3927–32.
Xu H, et al. A single amino acid substitution in human APOBEC3G antiretroviral enzyme confers resistance to HIV-1 virion infectivity factor-induced depletion. Proc Natl Acad Sci U S A. 2004;101(15):5652–7.
Anderson J, et al. Safety and efficacy of a lentiviral vector containing three anti-HIV genes--CCR5 ribozyme, tat-rev siRNA, and TAR decoy--in SCID-hu mouse-derived T cells. Mol Ther. 2007;15(6):1182–8.
Kumar P, et al. T cell-specific siRNA delivery suppresses HIV-1 infection in humanized mice. Cell. 2008;134(4):577–86.
ter Brake O, et al. Evaluation of safety and efficacy of RNAi against HIV-1 in the human immune system (Rag-2(−/−)gammac(−/−)) mouse model. Gene Ther. 2009;16(1):148–53.
Novina CD, et al. siRNA-directed inhibition of HIV-1 infection. Nat Med. 2002;8(7):681–6.
Kohn DB, et al. A clinical trial of retroviral-mediated transfer of a rev-responsive element decoy gene into CD34(+) cells from the bone marrow of human immunodeficiency virus-1-infected children. Blood. 1999;94(1):368–71.
Humeau LM, et al. Efficient lentiviral vector-mediated control of HIV-1 replication in CD4 lymphocytes from diverse HIV+ infected patients grouped according to CD4 count and viral load. Mol Ther. 2004;9(6):902–13.
Michienzi A, et al. A nucleolar TAR decoy inhibitor of HIV-1 replication. Proc Natl Acad Sci U S A. 2002;99(22):14047–52.
Zahn RC, et al. Efficient entry inhibition of human and nonhuman primate immunodeficiency virus by cell surface-expressed gp41-derived peptides. Gene Ther. 2008;15(17):1210–22.
Voit RA, et al. Generation of an HIV resistant T-cell line by targeted “stacking” of restriction factors. Mol Ther. 2013;21(4):786–95.
Lombardo A, et al. Gene editing in human stem cells using zinc finger nucleases and integrase-defective lentiviral vector delivery. Nat Biotechnol. 2007;25(11):1298–306.
Lombardo A, et al. Site-specific integration and tailoring of cassette design for sustainable gene transfer. Nat Methods. 2011;8(10):861–9.
Genovese P, et al. Targeted genome editing in human repopulating haematopoietic stem cells. Nature. 2014;510(7504):235–40.
Hoban MD, et al. Correction of the sickle cell disease mutation in human hematopoietic stem/progenitor cells. Blood. 2015;125(17):2597–604.
Wang J, Exline CM, et al. Highly efficient homology-driven genome editing in CD34+ hematopoietic stem/progenitor cells by combining zinc finger nuclease mRNA and AAV donor delivery. Nat Biotechnol. 2015;33(12):1256–1263.
Chu VT, et al. Increasing the efficiency of homology-directed repair for CRISPR-Cas9-induced precise gene editing in mammalian cells. Nat Biotechnol. 2015;33(5):543–8.
Maruyama T, et al. Increasing the efficiency of precise genome editing with CRISPR-Cas9 by inhibition of nonhomologous end joining. Nat Biotechnol. 2015;33(5):538–42.
Ye L, et al. Seamless modification of wild-type induced pluripotent stem cells to the natural CCR5Delta32 mutation confers resistance to HIV infection. Proc Natl Acad Sci U S A. 2014;111(26):9591–6.
Yao Y, et al. Generation of CD34+ cells from CCR5-disrupted human embryonic and induced pluripotent stem cells. Hum Gene Ther. 2012;23(2):238–42.
Chomont N, et al. HIV reservoir size and persistence are driven by T cell survival and homeostatic proliferation. Nat Med. 2009;15(8):893–900.
Chun TW, et al. Quantification of latent tissue reservoirs and total body viral load in HIV-1 infection. Nature. 1997;387(6629):183–8.
Finzi D, et al. Latent infection of CD4+ T cells provides a mechanism for lifelong persistence of HIV-1, even in patients on effective combination therapy. Nat Med. 1999;5(5):512–7.
Sarkar I, et al. HIV-1 proviral DNA excision using an evolved recombinase. Science. 2007;316(5833):1912–5.
Qu X, et al. Zinc-finger-nucleases mediate specific and efficient excision of HIV-1 proviral DNA from infected and latently infected human T cells. Nucleic Acids Res. 2013;41(16):7771–82.
Hauber I, et al. Highly significant antiviral activity of HIV-1 LTR-specific tre-recombinase in humanized mice. PLoS Pathog. 2013;9(9), e1003587.
Ebina H, et al. Harnessing the CRISPR/Cas9 system to disrupt latent HIV-1 provirus. Sci Rep. 2013;3:2510.
Blackard JT, et al. Transmission of human immunodeficiency type 1 viruses with intersubtype recombinant long terminal repeat sequences. Virology. 1999;254(2):220–5.
Surendranath V, et al. SeLOX—a locus of recombination site search tool for the detection and directed evolution of site-specific recombination systems. Nucleic Acids Res. 2010;38(Web Server issue):W293–8.
Karpinski J, et al. Universal Tre (uTre) recombinase specifically targets the majority of HIV-1 isolates. J Int AIDS Soc. 2014;17(4 Suppl 3):19706.
Ebina H, et al. A high excision potential of TALENs for integrated DNA of HIV-based lentiviral vector. PLoS One. 2015;10(3), e0120047.
Cong L, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23.
Zhu W, et al. The CRISPR/Cas9 system inactivates latent HIV-1 proviral DNA. Retrovirology. 2015;12:22.
Liao HK, et al. Use of the CRISPR/Cas9 system as an intracellular defense against HIV-1 infection in human cells. Nat Commun. 2015;6.
Hu W, et al. RNA-directed gene editing specifically eradicates latent and prevents new HIV-1 infection. Proc Natl Acad Sci U S A. 2014;111(31):11461–6.
Li H, et al. In vivo genome editing restores haemostasis in a mouse model of haemophilia. Nature. 2011;475(7355):217–21.
Joglekar AV, et al. Integrase-defective lentiviral vectors as a delivery platform for targeted modification of adenosine deaminase locus. Mol Ther. 2013;21(9):1705–17.
Holkers M, et al. Differential integrity of TALE nuclease genes following adenoviral and lentiviral vector gene transfer into human cells. Nucleic Acids Res. 2013;41(5), e63.
Chen Z, et al. Receptor-mediated delivery of engineered nucleases for genome modification. Nucleic Acids Res. 2013;41(19), e182.
Mock U, et al. Novel lentiviral vectors with mutated reverse transcriptase for mRNA delivery of TALE nucleases. Sci Rep. 2014;4:6409.
Khatri N, et al. In vivo delivery aspects of miRNA, shRNA and siRNA. Crit Rev Ther Drug Carrier Syst. 2012;29(6):487–527.
Morizono K, et al. Lentiviral vector retargeting to P-glycoprotein on metastatic melanoma through intravenous injection. Nat Med. 2005;11(3):346–52.
Lin AH, et al. Receptor-specific targeting mediated by the coexpression of a targeted murine leukemia virus envelope protein and a binding-defective influenza hemagglutinin protein. Hum Gene Ther. 2001;12(4):323–32.
Frecha C, et al. A novel lentiviral vector targets gene transfer into human hematopoietic stem cells in marrow from patients with bone marrow failure syndrome and in vivo in humanized mice. Blood. 2012;119(5):1139–50.
Anliker B, et al. Specific gene transfer to neurons, endothelial cells and hematopoietic progenitors with lentiviral vectors. Nat Methods. 2010;7(11):929–35.
Paraskevakou G, et al. Epidermal growth factor receptor (EGFR)-retargeted measles virus strains effectively target EGFR- or EGFRvIII expressing gliomas. Mol Ther. 2007;15(4):677–86.
Kneissl S, et al. Measles virus glycoprotein-based lentiviral targeting vectors that avoid neutralizing antibodies. PLoS One. 2012;7(10), e46667.
Kneissl S, et al. CD19 and CD20 targeted vectors induce minimal activation of resting B lymphocytes. PLoS One. 2013;8(11), e79047.
Alwin S, et al. Custom zinc-finger nucleases for use in human cells. Mol Ther. 2005;12(4):610–7.
Kim HJ, et al. Targeted genome editing in human cells with zinc finger nucleases constructed via modular assembly. Genome Res. 2009;19(7):1279–88.
Koo T, Lee J, Kim JS. Measuring and reducing off-target activities of programmable nucleases including CRISPR-Cas9. Mol Cells. 2015;38(6):475–81.
Hendel A, et al. Quantifying on- and off-target genome editing. Trends Biotechnol. 2015;33(2):132–40.
Pattanayak V, et al. Revealing off-target cleavage specificities of zinc-finger nucleases by in vitro selection. Nat Methods. 2011;8(9):765–70.
Gabriel R, et al. An unbiased genome-wide analysis of zinc-finger nuclease specificity. Nat Biotechnol. 2011;29(9):816–23.
Chiarle R, et al. Genome-wide translocation sequencing reveals mechanisms of chromosome breaks and rearrangements in B cells. Cell. 2011;147(1):107–19.
Frock RL, et al. Genome-wide detection of DNA double-stranded breaks induced by engineered nucleases. Nat Biotechnol. 2015;33(2):179–86.
Tsai SQ, et al. GUIDE-seq enables genome-wide profiling of off-target cleavage by CRISPR-Cas nucleases. Nat Biotechnol. 2015;33(2):187–97.
Crosetto N, et al. Nucleotide-resolution DNA double-strand break mapping by next-generation sequencing. Nat Methods. 2013;10(4):361–5.
Ran FA, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–98.
Kim D, et al. Digenome-seq: genome-wide profiling of CRISPR-Cas9 off-target effects in human cells. Nat Methods. 2015;12(3):237–43, 1 p following 243.
Doyon Y, et al. Enhancing zinc-finger-nuclease activity with improved obligate heterodimeric architectures. Nat Methods. 2011;8(1):74–9.
Tsai SQ, et al. Dimeric CRISPR RNA-guided FokI nucleases for highly specific genome editing. Nat Biotechnol. 2014;32(6):569–76.
Acknowledgements
We gratefully acknowledge the expertise of our collaborators at Sangamo BioSciences, including Michael Holmes, Jianbin Wang and Philip Gregory, and thank Liz Wolffe for her help compiling Table 1. This work was supported by the James B. Pendleton Charitable Trust, NIH grants HL073104, AI110149 and HL129902, and the California HIV/AIDS Research Program grant ID12-USC-245.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 American Society of Gene and Cell Therapy
About this chapter
Cite this chapter
Llewellyn, G.N., Exline, C.M., Holt, N., Cannon, P.M. (2016). Using Engineered Nucleases to Create HIV-Resistant Cells. In: Cathomen, T., Hirsch, M., Porteus, M. (eds) Genome Editing. Advances in Experimental Medicine and Biology(). Springer, New York, NY. https://doi.org/10.1007/978-1-4939-3509-3_10
Download citation
DOI: https://doi.org/10.1007/978-1-4939-3509-3_10
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-3507-9
Online ISBN: 978-1-4939-3509-3
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)