Abstract
From the moment a developing thymocyte expresses a TCR, it is subjected to numerous interactions with self-peptide/MHC complexes that determine its ultimate fate. These include death by neglect, negative selection (apoptosis and lineage deviation), positive selection, and lineage commitment. The identification of signals that govern these unique cell fates requires the ability to assess the activity, level of expression, subcellular location, and the molecular associations of numerous proteins within the developing T cell. Thus, this chapter describes methods designed to analyze thymocyte signaling under various types of peptide-based stimulation in vitro.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Key words
1 Introduction
The selection of functional T cells is mediated by interactions between the T cell antigen receptor and self-peptide/major histocompatibility complex expressed on thymic epithelium. These interactions either lead to survival and development, or death. The affinity model proposes that the selection outcome is determined by the affinity of the TCR for a peptide–MHC complex. Weak TCR/peptide–MHC interactions do not support thymocyte survival (death by neglect); strong interactions lead to thymocyte apoptosis, lineage deviation or receptor editing (collectively called negative selection); and interactions between these extremes lead to the development of mature T cells (positive selection) [1]. More recently, a number of “third” signals have been given a role in the selection process [2]. A longstanding issue remains as to how the TCR reads the parameters of ligand engagement, along with these third signals, to direct these distinct cell fates.
We use several standard biochemical techniques to assess signals in our system. Immunoblot techniques are used to determine the expression levels and activation states of proteins in large synchronous populations of thymocytes: for these assays we stimulate, lyse, SDS-PAGE, transfer and immunoblot (see below for stimulation and lysis protocols). Flow cytometry is used to perform similar assays on small/rare populations of cells that can be distinguished by a specific phenotypic marker (also works for large populations): for these assays we stimulate, stain surface markers and perform intracellular staining. In general we use standard kits (e.g. BD Phosflow or Cytofix/Cytoperm kits) and the methods are not described here. However, we have used the fixation and permeabilization protocol reagents below in single cell suspension to assess expression levels of a number of different proteins with some success. Confocal microscopy provides activation and location information that has added to our understanding of the diversity in the regulation of selection signals [3, 4]. These assays are described below.
Historically, immunoprecipitation followed by western blotting (IP-Western) has been invaluable for elucidating the protein–protein interactions that occur during thymocyte positive and negative selection in the thymus. We have used these assays extensively to map out differences in signaling in large synchronous populations of T cells available from thymi of mice on a non-selecting background [3]. However, IP-Western can have significant drawbacks when targeting small/rare populations of T cells, proteins that are expressed at low levels, or proteins whose available reagents are inefficient at IP. Thus, IP followed by flow cytometry (IP-FCM) offers the benefit of being able to precipitate low abundance proteins to generate data with a high signal to noise ratio [5–7]. Additionally, this protocol offers the benefit of being able to use significantly fewer cells for analysis (~1 × 105 for IP-FCM vs. 100 × 106 for IP-Western). Others have even successfully performed these types of analyses on <1 × 103 cells [7]. These assays are described below.
2 Materials
Prepare all solutions using autoclave sterilized MilliQ (or equivalent) water and molecular biology grade reagents. All reagents are stored at room temperature unless otherwise noted.
2.1 Tissue Preparation
-
1.
Complete media: RPMI1640 supplemented with 1 mM non-essential amino acids (NEAA), 1 mM sodium pyruvate, 50 μM β-Mercaptoethanol, 100 U/mL Penicillin 100 μg/mL Streptomycin 0.292 mg/mL Glutamine, and 10 % FBS.
-
2.
TCR transgenic mouse on a non-selecting background. For CD8 T cell selection TCR transgenic Rag−/− β2m−/− mouse preferably <6 weeks of age are ideal (see Note 1 ).
-
3.
Alternatively, to assess cohorts of T cells at multiple stages of development, thymi from TCR transgenic and polyclonal mice on a selecting background can also be used (see Note 2 ).
2.2 Stimulation with Peptide-Pulsed APC (See Note 3 )
-
1.
Antigen-presenting cells: examples: bulk splenocytes, T cell-depleted spleens, bone marrow-derived dendritic cells or other cell lines that express the appropriate MHC.
-
2.
Peptide solution, concentration previously optimized.
2.3 Stimulation with Peptide/MHC I Tetramers
-
1.
Peptide/MHC Tetramers (2 mg/mL).
-
2.
Complete media (see Subheading 2.1).
2.4 Confocal Microscopy
-
1.
13 mm round glass coverslips (no. 1: 0.13 mm thick).
-
2.
Microscope slides (pre-cleaned) 25 × 75 × 1.0 mm.
-
3.
Humidity chamber: 6-well plates with a coverslip on diameter filter paper, parafilm and ~1 mL water (Fig. 1) (see Note 4 ).
Fig. 1 Schematic of a humidity chamber for confocal microscopy. In general we prepare a 6-well plate with a filter paper, covered by piece of parafilm with approximately 1 mL of water. The filter paper and parafilm are circles that we mark and cut using the diameter of a 50 mL conical tube as a template. The cover of the plate is labeled with the conditions (stimulation, stain, time, etc.). These are easily reused from experiment to experiment
-
4.
Complete media (see Subheading 2.1).
-
5.
0.2 % Triton solution.
-
6.
Fresh 1.5 and 3.0 % (w/v) BSA/PBS solution.
-
7.
Staining conditions for surface and/or cytosolic proteins: prepare a stock solution of 4 % formaldehyde fix (1 part 16 % formaldehyde plus 3 parts PBS). Make and keep at RT in chemical hood.
-
8.
Staining conditions for cytosolic and/or nuclear proteins: prepare a stock solution of 8 % formaldehyde fix (1 part 16 % formaldehyde plus 1 part PBS). Make and keep at RT in chemical hood.
-
9.
Blocking buffer: 25 mL 3 % (w/v) BSA/PBS. Keep on ice until use.
-
10.
OPTIONAL: DNA/nuclear staining reagent such as DAPI or DRAQ-5AT if nuclear localization is desirable.
2.5 IP-FCM
-
1.
Lysis Buffer: 10 mM Tris pH 7.4, 1 % Triton X-100, 0.5 % NP-40, 150 mM NaCl, 1 mM EDTA, 1 mM EGTA. Add 20 mM NaF, 0.2 NaVO4, and 0.2 mM PMSF fresh every time before use.
-
2.
Carboxylate-modified polystyrene latex (CML) beads ~6 μM diameter (Store at 4 °C).
-
3.
Monoclonal antibodies for IP in PBS (≥0.2 mg/mL, must be devoid of BSA).
-
4.
Antibodies for detection (Must be directly conjugated to a fluorochrome or a species other than IP mAb to avoid cross-reactivity).
-
5.
MES Coupling Buffer: 50 mM 2-(N-morpholino)ethanesulfonic acid (MES) pH 6.0, 1 mM EDTA.
-
6.
EDAC-MES: 50 mg/mL 1-ethyl-3-(3-dimethylaminopropyl)carbodiimide hydrochloride (EDAC).
-
7.
Quenching/Blocking/Storage (QBS) Buffer: 1 % BSA + 0.02 % Sodium azide in 1× PBS (Store at 4 °C).
-
8.
FCM Staining Buffer: 1 % FBS + 0.2 % Sodium azide in 1× PBS (Store at 4 °C).
3 Methods
3.1 Harvesting Thymi (See Note 5 )
-
1.
Prepare sterile tools and small Petri dishes with 1 mL of complete media and 1 piece of sterile nylon mesh.
-
2.
Place Petri dish on ice.
-
3.
Harvest thymi from mice that have been euthanized according to your institutional guidelines. Place in the Petri dish on ice (one thymus per Petri dish).
-
4.
Place another piece of mesh over the thymus and press with a plunger from a 3 mL syringe to create a single cell suspension.
-
5.
Pass cells to a clean 15 mL conical tube that contains 3 mL of complete media.
-
6.
Rinse the mesh with 1 mL of fresh complete media and transfer to the conical tube.
-
7.
Spin cells at 483 × g (450–500 × g) for 5 min in centrifuge (4 °C).
-
8.
Resuspend cells in fresh complete media (1–2 mL/thymus).
-
9.
Transfer the cells to a new conical (50 mL) through cell strainer (or nylon mesh) to remove large clumps of cells and thymic debris.
-
10.
Count with a hemocytometer (or electronic counter) and dilute cells to a concentration 10 × 106/mL. Proceed to stimulation of your choice.
3.2 Stimulation with APC
3.2.1 Peptide-Loaded APC
-
1.
Prepare media that contains the peptide at the desired concentration (see Note 6 ).
-
2.
Resuspend APC at 20 × 106/mL in the peptide solution (from step 1).
-
3.
Incubate at 37 °C in the culture incubator from a minimum of 30 min.
-
4.
Wash cells with complete media, spin at 483 × g (450–500 × g) for 5 min and discard supernatant.
-
5.
Resuspend cells at 10 × 106 cells/mL of complete media.
3.2.2 Stimulation with Peptide-Loaded APC
-
1.
Set the thermomixer or water bath at 37 °C.
-
2.
Mix T cells and antigen-loaded APCs at a ratio 1:1. Ideally, for confocal microscopy or ICS-flow cytometry, mix 50 μL APCs (10 × 106/mL) with 50 μL T cells (10 × 106/mL) in a 1.5 mL microfuge tube.
-
3.
Centrifuge in bench top mini-centrifuge. Use pulse/quick spin mode for 7–10 s.
-
4.
Tap the pellet.
-
5.
Place in thermomixer at 37 °C for the planned time course.
-
6.
Place the tubes on ice to stop stimulation.
3.2.3 Stimulation with Peptide/MHC I Tetramers
-
1.
Add approximately 1 × 106/staining condition to 1.5 mL microfuge tube. Example: for an experiment requiring three staining conditions for each time point, one would add 3 × 106 thymocytes (300 μL of 10 × 106/mL suspension of thymocytes from above).
-
2.
Place cells in thermomixer (37 °C, 200–500 rpm mixing).
-
3.
Add desired concentration of peptide/MHC tetramers and incubate for planned time course (see Note 7 ).
-
4.
Wash 2× with ice-cold media (to remove excess tetramer) and place the tubes on ice to stop stimulation.
3.3 Confocal Microscopy
3.3.1 Mounting Cells
-
1.
Wash cells from stimulation (above) and resuspend pellet (10 × 106 cells/mL) by gently tapping the microfuge tube (see Note 8 ).
-
2.
Place 100 μL of stimulated cells on the appropriate coverslip. TAKE NOTE OF PLATE LAYOUT.
-
3.
To allow the cells to settle onto the coverslip, carefully place covered 6-well plates in refrigerator for minimum of 10 min. During this time prepare the staining buffers. Fixation: (see Note 9 ).
-
4.
Move plates to chemical hood and add:
-
(a)
For surface and/or cytosolic stains: 100 μL of 4 % fix. Incubate at RT for 45 min.
-
(b)
For cytosolic and/or nuclear stains: 100 μL of 8 % fix. Incubate at RT for 30 min.
-
(a)
-
5.
Carefully aspirate liquid off each coverslip and wash 2× with 200 μL PBS. (You may stop the procedure at this step. To do this, add 200 μL PBS and place plates at 4 °C). For cytosolic and nuclear stains proceed to step 10.
3.3.2 Permeablization/Blocking
-
1.
Gently, but quickly add 100 μL of ice cold 0.2 % triton drop-wise and incubate for 2 min. (For the best results, work with six wells at a time. Start the timer after adding the triton to last coverslip.)
-
2.
Aspirate triton and add ice cold 3 % BSA/PBS.
-
3.
Repeat steps 5 and 6 until all coverslips have been treated.
-
4.
Place plates at 4 °C overnight. Alternatively, block for a minimum of 1 h then proceed to staining procedure.
3.3.3 Staining
-
1.
Prepare staining cocktails.
-
(a)
For primary surface and cytosolic stains, add the desired antibodies to 1.5 % BSA/PBS (w/v).
-
(b)
For primary cytosolic and nuclear stains, add desired antibodies to 1.5 % BSA/0.2%Triton solution.
-
(c)
Prepare secondary antibodies by adding labeled antibodies to 1.5 % BSA/PBS (w/v).
-
(a)
-
2.
Aspirate coverslip and wash 2× with RT PBS. For cytosolic and nuclear stains skip ahead to step 16 .
-
3.
Add primary antibody staining cocktail. TAKE NOTE OF PLATE LAYOUT!!!
-
4.
Incubate at RT in the dark for 1 h. During the staining incubations make labels for slides.
-
5.
Wash 2× with PBS and add secondary staining cocktail.
-
6.
Incubate at RT in the dark for 1 h. For surface and cytosolic stains proceed to step 17 .
-
7.
Add 50 μL of the primary stain cocktail in PBS/BSA 1.5 %/Triton 0.2 % for 30 min at RT
-
8.
Wash three times in PBS/BSA1.5 %
-
9.
Add secondary staining cocktail and incubate for 1 h.
-
10.
OPTIONAL: If DNA/Nuclear staining is desired, dilute DRAQ-5 at 1/5000 (or other DNA stain) in PBS/formaldehyde 1 %. Add 50 μL to the coverslip and incubate for 5 min at RT.
3.3.4 Mounting the Coverslip
-
1.
Using a forceps to pick up coverslip, dab the edge of the coverslip with a lint-free tissue to remove excess stain. Then dip the coverslip in PBS and dab the edge of the coverslip with tissue three times, then dip in water and dab once. Place the coverslip CELL SIDE DOWN, onto a microscope slide that has one drop of mounting media (see Note 10 ).
-
2.
Cover the mounted coverslip/slides and let dry overnight at RT, then paint the edges of each coverslip with clear nail polish (to make a seal) and place in slide box and place at 4 °C. NOTE: The slides are then ready for analysis by confocal microscopy. While in theory the stains remain stable for up to a year, we suggest they be analyzed within 3 months of mounting.
3.4 IP-FCM
3.4.1 Generation of Ab-Coupled CML Beads
-
1.
Determine the concentration of CML beads by diluting beads in PBS and enumerating using a hemacytometer under a microscope.
-
2.
Gently vortex CML beads to suspend them evenly and transfer 20 × 106 beads to a 1.5 mL centrifuge tube.
-
3.
Wash the beads three times in 0.5 mL MES Coupling Buffer, centrifuging at ~16,000 × g for 5 min at 25 °C.
-
4.
Following last wash, aspirate buffer leaving beads undisturbed. Resuspend beads in 50 μL MES Coupling Buffer.
-
5.
Add 20 μL EDAC-MES (made fresh) and mix by pipetting up and down for 15 min to prevent beads from settling.
-
6.
Wash the beads three times in 0.5 mL MES Coupling Buffer, centrifuging at ~16,000 × g for 5 min at 25 °C.
-
7.
Aspirate buffer leaving beads undisturbed and resuspend beads in 50 μL PBS.
-
8.
Add 50 μL mAb of choice.
-
9.
Mix for 3–4 h in a thermomixer at ~1100 rpm at 25 °C, being sure that no beads are settling at the bottom of the tube.
-
10.
Wash beads three times in 1 mL PBS, centrifuging at ~16,000 × g for 5 min at 25 °C being sure to discard as much supernatant as possible without disturbing the CML bead pellet.
-
11.
Following last wash, aspirate all PBS and resuspend beads in 100 μL QBS Buffer and store overnight at 4 °C.
-
12.
Count beads by diluting in PBS and enumerating using a hemacytometer under a microscope.
3.4.2 Sample Lysis
-
1.
Lyse 10 × 106 thymocytes in 100 μL Lysis Buffer (ice-cold) in a 1.5 mL microcentrifuge tube for 20 min on ice.
-
2.
Following lysis spin crude lysate for 50 min at ~16,000 × g at 4 °C.
-
3.
Pass supernatant to a fresh 1.5 mL microcentrifuge tube (see Note 11 ).
-
4.
Add 2.5 × 105 IP beads to the lysate (see Note12).
-
5.
Place tubes in a thermomixer at ~1100 rpm overnight (place mixer itself in a 4 °C fridge), making sure that beads do not settle.
3.4.3 Staining of Beads for FCM
-
1.
Wash CML beads that have been incubated with lysate three times in ice-cold Lysis Buffer being sure to discard as much buffer as possible without disturbing the CML bead pellet.
-
2.
Resuspend CML beads in FCM staining buffer and pass beads to 96-well round-bottom plate.
-
(a)
Split the CML beads into enough wells to assess the proteins of interest plus isotype control(s).
-
(a)
-
3.
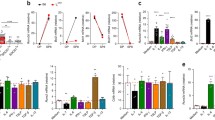
Add fluorochrome-conjugated mAbs (or primary Abs) to the samples and incubate for 30 min on ice in darkness. α-POSH, clone M-290, α-JIP-1 clone M-300 and α-JNK1 clone C-17 used for demonstration (Fig. 2) [5].
Fig. 2 IP-FCM analysis of protein interactions in pre-selection DP thymocytes. (a) Gating on singlet beads based on FSC and SSC properties. (b) Proper calibration of fluorescence voltage such that the isotype control falls between 100 and 101 while a “bright” positive control falls on scale. (c) Representative experiment where α-POSH CML beads are used on lysates of pre-selection DP thymocytes. Histograms represent the presence of JIP-1 and JNK1 bound to POSH. Red histogram represents the “loading control” to quantify how much POSH was precipitated
-
4.
Wash beads two times with FCM Buffer, spinning at ~500 × g for 5 min at 25 °C.
-
5.
If unlabeled primary Abs are used, add fluorochrome-conjugated secondary Ab and incubate for 30 min on ice in darkness. If directly conjugated Abs are used, skip this step.
-
6.
Resuspend IP beads in 200 μL FCM Staining Buffer and proceed to analysis on the flow cytometer.
3.4.4 Acquisition of Data by FCM
-
1.
Increase the FSC and SSC of the cytometer such that bead singlets are clearly visible (see Note 13 ).
-
2.
Draw a gate around the population of bead singlets (Fig. 2a).
-
3.
Apply this gate and visualize the fluorescence data on a log scale for each of the fluorescence channels being used.
-
4.
Pass the isotype control sample and confirm that the generated histogram falls between 100 and 101 (see Note 14 ).
-
5.
Pass the control for a very bright sample (see Note 15 ).
-
6.
Pass experimental samples (Fig. 2c).
- 7.
4 Notes
-
1.
Older mice will work. However, they have significantly fewer pre-selected DP.
-
2.
Due to the heterogeneity of the T cell populations from these thymi, confocal and flow cytometry-based assays are more suitable to assess signaling in these T cells.
-
3.
Due to potential contamination with APC-derived proteins, this method is not appropriate for immunoblotting, IP or IP-FCM.
-
4.
Carefully plan out the stains and time course in advance. Label the cover of the 6-well plate with the stimulation, stain and timepoint for each coverslip. Keep in mind how you want to collect the data at the confocal. One slide holds three coverslips max. An intelligent and logical layout greatly simplifies the collection and analysis of the data. In addition, it greatly reduces the probability of mislabeled coverslips.
-
5.
For short-term assays, those that are performed in less than 24 h, this step can be performed at the bench. Longer time points require FTOC (described in Chapter 12) and should be performed in a sterile hood.
-
6.
When using peptides of different strengths, always start by handling the weakest peptide and move step by step to the strongest peptide.
-
7.
You may substitute anti-CD3 with or without cross-linking. When using cross-linking, pre-incubate thymocytes with anti-CD3 on ice for 15 min, wash, and add pre-warmed media with appropriate concentration of cross-linking antibody, start timing.
-
8.
For analyses involving confocal examination of thymocyte/APC conjugates do not mix cells by pipetting.
-
9.
If your cell surface target is sensitive to fixation, stain on ice after stimulation and prior to fixation).
-
10.
To avoid bubbles, hold coverslip cell side down at 45 % angle and slide the coverslip across slide until it touches the drop of mounting media then gently lay the coverslip down onto slide.
-
11.
Alternatively, one can proceed to standard SDS-PAGE/immunoblot (described elsewhere [3]) procedures at this point.
-
12.
The number of thymocytes used, the volume of lysis buffer, and the number of CML beads used must be determined empirically based on the proteins being analyzed.
-
13.
The CML beads being used are smaller than thymocytes, as such the FSC and SSC gain will have to be increased to make them visible to the cytometer.
-
14.
If fluorescence is too bright lower the channel voltage, if the histogram does not show, increase the channel voltage. α-Rabbit-488 used for demonstration purposes (see Fig. 2b).
-
15.
A good control for a very bright sample is anti-mouse Ig (if your Ab coupled to the CML bead is mouse) as it is highly abundant on the CML bead. α-Mouse 488 was used for demonstration purposes (see Fig. 2b).
References
Starr TK, Jameson SC, Hogquist KA (2003) Positive and negative selection of T cells. Annu Rev Immunol 21:139–176
Stritesky GL, Jameson SC, Hogquist KA (2012) Selection of self-reactive T cells in the thymus. Annual Rev Immunol 30:95–114
Daniels MA, Teixeiro E, Gill J, Hausmann B, Roubaty D, Holmberg K, Werlen G, Holländer GA, Gascoigne NR, Palmer E (2006) Thymic selection threshold defined by compartmentalization of Ras/MAPK signalling. Nature 444(7120):724–729
Daniels MA, Teixeiro E (2010) The persistence of T cell memory. Cell Mol Life Sci 67(17):2863–2878
Cunningham CA, Knudson KM, Peng BJ, Teixeiro E, Daniels MA (2013) The POSH/JIP-1 scaffold network regulates TCR-mediated JNK1 signals and effector function in CD8 T cells. Eur J Immunol 43(12):3361–3371
Gil D, Schrum A, Alarcon B, Palmer E (2005) T cell receptor engagement by peptide – MHC ligands induces a conformational change in the CD3 complex of thymocytes. J Exp Med 201(4):517–522
Schrum AG, Gil D, Dopfer EP, Wiest DL, Turka LA, Schamel WW, Palmer E (2007) High-sensitivity detection and quantitative analysis of native protein-protein interactions and multiprotein complexes by flow cytometry. Sci STKE 2007(389):pl2
Acknowledgements
The authors thank the members of the Daniels and Teixeiro labs for assistance in compiling these protocols. Funding provided by grants from the Missouri Mission Enhancement Fund and University of Missouri Research Board is greatly acknowledged.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer Science+Business Media New York
About this protocol
Cite this protocol
Cunningham, C.A., Teixeiro, E., Daniels, M.A. (2016). In Vitro Analysis of Thymocyte Signaling. In: Bosselut, R., S. Vacchio, M. (eds) T-Cell Development. Methods in Molecular Biology, vol 1323. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-2809-5_15
Download citation
DOI: https://doi.org/10.1007/978-1-4939-2809-5_15
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-2808-8
Online ISBN: 978-1-4939-2809-5
eBook Packages: Springer Protocols