Abstract
Bacterial motility is ubiquitous and essential to complex lifestyles including host infection and biofilm formation. While a tremendous amount of work has been dedicated to understand the mechanism of bacterial swimming in liquid with a rotary flagellum, much less is understood about mechanisms allowing bacteria to colonize surfaces. One such mechanism, called gliding motility, allows the smooth translocation of rod-shaped bacteria along their long axis in the absence of visible external organelles. While the mechanism of gliding has long been mysterious, recent work in two model systems, Myxococcus xanthus and Flavobacterium johnsoniae have shed new light on the propulsion mechanism. In both systems, key features of the motility processes were revealed by imaging the dynamics of critical motility proteins in real-time. Generally, these studies suggest that both Myxococcus and Flavobacterium appear to use dynamic helical envelope assemblage for their movement, but there are also important differences in the motility mechanisms. In this chapter, we review recent progress on the molecular characterization of the Myxococcus and Flavobacterium motility machineries. Beyond motility, this research will likely impact our understanding of dynamic processes in the bacterial cell envelope.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Secretion System
- Motility Mechanism
- Propulsion Mechanism
- Total Internal Reflection Fluorescence
- Total Internal Reflection Fluorescence Microscopy
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
In bacteria, motility is a ubiquitous phenotypic trait allowing a variety of lifestyles and environmental adaptations. Bacterial motility is pivotal to biofilm formation and can also support virulence. For these reasons, motility and its regulation have been intensively studied in a variety of bacterial model systems. However, while these studies have shed light to fundamental aspects of bacterial motility, they have largely focused on 3D swimming in viscous media using a flagellum (Jarrell and McBride 2008) . Yet, bacteria are also capable of moving over solid surfaces, which is important for a number of cooperative behaviors. One mode of surface locomotion, called twitching motility is relatively well understood and employs fibrillar appendages called type-IV pili (T4P) that pull cells like retractile grappling hooks (Skerker and Berg 2001) . Twitching motility is widespread in bacteria and although the molecular mechanisms underlying pilus function still need to be resolved, the propulsion mechanism is relatively unambiguous. Another form of surface motility, called gliding motility , occurs without the aid of pili, flagella, or any obvious organelles, and without observable changes in cell morphology (Jarrell and McBride 2008). Gliding motility is observed in very diverse phylogenetically unrelated bacterial groups and has been studied mechanistically in the Cyanobacteria, the Mollicutes, the Bacteroidetes, and the Myxobacteria (Mignot 2007) . Studies in these organisms suggest that in each case, the gliding motility mechanism is distinct and does not result from a universal gliding machinery. Thus, gliding motility may have evolved independently on several occasions and may involve more than one motility mechanism. While mechanistic studies are available in the Cyanobacteria, the lack of molecular tools has hampered in-depth characterization of the motility mechanism and currently, the cyanobacterial motility machinery remains elusive. Molecular work in the Mollicutes has shown that Mycoplasma can use a variety of mechanisms involving the so-called terminal organelle of Mycoplasma pneumoniae and large leg proteins located at the “neck” of Mycoplasma mobile (Mignot 2007) . Since the Mollicutes have a very particular cell architecture and their motility mechanisms are mostly likely unique and clade-specific, they are not discussed here (see Mignot 2007 for more information). In recent years, much progress has been made to understand gliding motility in the myxobacteria and in the Bacteroidetes. While the gliding mechanisms share common features, key differences are also observed. In this book chapter, we review the latest findings on each of these systems and attempt to delineate general principles underlying this complex and widespread form of bacterial locomotion.
6.1 Gliding Motility in Myxococcus Xanthus
Myxococcus xanthus, a member of the delta-subgroup of bacteria, displays a remarkable multicellular lifestyle . When facing starvation, Myxococcus enters a developmental program where thousands of Myxococcus cells cooperate to build multicellular structures called fruiting bodies, wherein the cells differentiate into environmentally resistant spores (Kaiser 2003) . To realize this life cycle, M. xanthus uses two distinct motility systems (Hodgkin and Kaiser 1979) . One motility system, the so-called social motility (S-motility), promoting the coordinated movement of large cell groups, consists of a polar T4P. S-motility is driven by the tight cooperation of T4P and a specific surface exopolysaccharide (EPS; Li et al. 2003) . S-motility is therefore a cooperative form of twitching motility and has been reviewed extensively (Zhang et al. 2012) . This book chapter discusses the second motility system, called the A-motility system. Myxococcus cells that lack T4P are still able to move as individual cells (hence the term adventurous (A)-motility), smoothly along their long axis by a typical gliding motility process. Because this movement occurs in absence of visible extracellular organelles (Burchard 1981) , its mechanism has been a mystery for a long time . One visible manifestation of A-motility is the deposition of a mucus (slime) , readily observable by a phase contrast microscopy of A-motile Myxococcus cells (Beebe 1941) . Since Cyanobacteria also deposit slime trails, which could be directly linked to the propulsion of cyanobacterial filaments (Dhahri et al. 2013; Hoiczyk and Baumeister 1998) , it has also been proposed that Myxococcus A-motility is driven by slime secretion through jet-like secretory organelles located at the back of the cells. However, the recent characterization of the A-motility machinery and its localization in live moving cells argues strongly against this model. Moreover, slime secretion has been observed at high resolution which suggests a function in adhesion (see below) .
6.1.1 The Myxococcus A-Motility Machinery
The A-Motility Machinery Forms at Bacterial Focal Adhesion
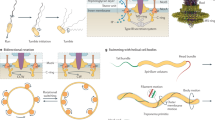
The mechanism of A-motility was suggested in 2007 by single cell studies of a critical A-motility protein, AglZ. Using a strain expressing a functional AglZ–YFP fusion protein, Mignot et al. (2007) observed that AglZ localized in clusters, distributed at regular intervals along the cell body (Figure 6.1a). Remarkably, time-lapse experiments revealed that these clusters were initially assembled at the leading pole and retained a fixed position with respect of the substratum as the cell moved forward. Since the cell was in motion, the clusters appeared stationary relative to the substratum (hence a fixed reference), but they were in fact moving in the direction opposite to the direction of movement and at the same velocity. The clusters eventually reached the back of the cell where they became dispersed (Figure 6.1a). Based on these observations, Mignot et al. (2007) proposed that the AglZ clusters reflect the localization of the A-motility machinery. This machinery would consist of intracellular motors moving on cytoplasmic cytoskeletal filaments and transmitting force through the cell envelope , which ultimately would cause the forward translocation of the cell body (Figure 6.1b). This hypothesis predicted that the motility machinery localizes at the focal adhesion sites .
The focal adhesion hypothesis. a AglZ–YFP localizes to periodic sites that remain fixed relative to the substratum in a moving cell. Overlay of the phase and the YFP (magenta, artificially colored for improved clarity) images captured every 30 s are shown. Arrowheads highlight selected bright fluorescence clusters. Scale bar = 2 μm (modified from Mignot 2007). b Motility mechanism suggested by Mignot et al. (2007). In this model, large focal adhesion complexes would penetrate the cell envelope, stick to the substratum at one end, and connect to cytoskeletal filaments at the other end. Cytoskeletal motor proteins would travel opposite to the direction of the cell and thus push backward (small arrows) against the adhesions, thus propelling the cell forward. (Modified from Nan and Zusman (2011)
Identification of the A-Motility Machinery
Although the AglZ protein might point to the localization of the motility machinery, it is not per se a structural component of this machinery because genetic evidence suggests that it functions to regulate its spatial positioning (Luciano et al. (2011; Mauriello et al. (2009) . The structural components of the machinery itself have long remained elusive. In fact, the genetic evidence for the existence of A-motility was obtained as early as in 1979 by Hodgkin and Kaiser, who found that two distinct sets of genes promote Myxococcus motility independently (Hodgkin and Kaiser (1979) . However, while the first set of genes was found to encode a T4P (the S-motility apparatus), the molecular identity of the A-motility system remained unknown until very recently. Over the years, several genetic screens have sought to identify the A-motility machinery; however, while approximately 50 genes were identified, their annotation did not reveal a conspicuous molecular machinery, mainly because the inactivation of many housekeeping genes is also likely to impair motility (Youderian et al. (2003; Yu and Kaiser (2007) . In addition, transposons were found in many genes of unknown function, which also rendered the identification of a gliding machinery difficult (Youderian et al. 2003; Yu and Kaiser 2007) .
To identify the gliding machinery proteins, 51 genes previously identified by transposon-based genetic screens (Youderian et al. 2003; Yu and Kaiser 2007) were reinvestigated under the assumption that if some of them encoded the actual machinery components, they must have coevolved. Doing so, three main genetic loci, a seven-gene operon (named gltD-J), a four-gene operon (gltA-C and gltK), and a three-gene operon (aglR-S; Figure 6.2a), for a total of 14 genes, became apparent (Luciano et al. 2011) . Remarkably, the taxonomic distribution of these genes revealed that 7 of the 14 genes were clustered together in several bacterial genomes and this core set of genes contained genes of each of the three separate loci identified in Myxococcus, suggesting that they encode a functional machinery (Luciano et al. 2011) . These findings suggested that the Myxococcus A-motility machinery may consist of up to 14 genes and could have emerged by the functional specialization of a broadly conserved core system of seven genes. The predicted function of the agl and glt genes also suggested that they encode a transenvelope complex (Figure 6.2b). Remarkably, the agl genes encode a flagellar stator homologue, a class of ion-driven motor proteins. The direct function of the agl and glt genes in A-motility was further proven experimentally (Luciano et al. 2011 , see below) and thus the machinery was named Agl–Glt. An independent biochemical search of the A-motility machinery identified critical interactions between some of the Glt proteins as essential for A-motility, confirming that the A-motility machinery had been identified (Nan et al. 2010) .
The Agl–Glt complex. a Genetic organization of the 14 genes encoding the components of the gliding machinery in Myxococcus xanthus. The G1 and G2 clusters correspond to the glt genes, and M1 cluster corresponds to the aglRQS genes. b Predicted structures of the Agl–Glt machinery based on experimental and bioinformatics predictions. The PG is not represented because its connection to Glt proteins is unknown
When possible, functional fusions were made to the components of the Agl–Glt complex. All tested proteins (GltD, GltF, AglQ, and AglR) localized to the focal adhesion sites together with AglZ, showing unambiguously that the Myxococcus A-motility machinery is assembled at these sites (Luciano et al. 2011; Nan et al. 2010) .
The Motility Motor is a Flagellar Stator Homologue
The agl genes encode proteins with similarities to the flagellar stator proteins (MotAB) or the TolQR/ExbBD proteins that energize colicin and iron siderophore transport, respectively (Sun et al. 2011) . This class of bacterial proton-conducting channels operates by harnessing the proton gradient across the inner membrane to generate mechanical force (Cascales et al. 2000) . Consistent with proton motive force (PMF) acting as the major energy source for A-motility, PMF uncoupling drugs (carbonyl cyanide m-chlorophenylhydrazone (CCCP)) and more specifically pH gradient-dissipating drugs (Nigericin), rapidly and reversibly inhibited A-motility (Sun et al. 2011) . In frame deletions of the algRQS genes further led to a complete defect in gliding motility . The Agl complex is predicted to form a complex where AglR associates both with AglQ and AglS to form a transmembrane proton channel (Figure 6.2b). Consistent with this, the mutation of a conserved aspartate in AglQ (D28N) predicted to bind H+ ions in the lumen of the channel, abolished A-motility (Sun et al. 2011) . Importantly, the motility complex is still assembled at focal adhesions sites in the AglQD28N mutant but it is not dynamics, further suggesting that the Agl complex is not involved in the assembly of the complex but in its energization. Last, the localization of AglQ-mCherry overlapped with that of AglZ–YFP at the focal adhesion complexes (FAC), proving that focal adhesions contain the energy-producing component of the motility machinery (Figure 6.3a; Sun et al. 2011) . How exactly Agl motor activity translates into motion remains to be understood, but based on knowledge of TolQR-TolA studies, it is currently thought that a conformational change in the lumen of the Agl channel is transduced to the TolA-like GltG protein and ultimately to the bacterial outer membrane. Consistent with this, AglR has been shown to interact directly with GltG (Luciano et al. 2011) . Force-generation by the Agl complex was shown directly by adding polystyrene beads to the outer surface of cells with an optical trap. When the beads collided with traveling complexes, they became bound and co-tracked with these complexes directionally towards the lagging cell pole. Experiments with agl mutants and PMF uncouplers proved that bead-transport was energized by the Agl complex (Sun et al. 2011) .
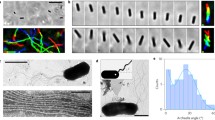
Localization of Myxococcus motility proteins. a Time lapse of a cell expressing AglQ-mCherry is shown. Fixed clusters are marked by white arrowheads. Fluorescent micrographs were taken every 15 s. Scale bar = 1 μm (reprinted with permission from Ducret et al. 2012). b Slime patches are deposited where the Agl/Glt machinery assembles. Slime was stained with a lectin after the cell left the positions shown on A and B. Triangular arrows point to fixed AglQ-bright motility complexes at positions where conspicuous slime patches were deposited. Scale bar = 1 μm (reprinted with permission from Ducret et al. 2012). c Deconvolved images of potential gltD-mCherry helices. Scale bar = 1 μm. (reprinted with permission from Nan et al. 2011)
6.1.2 The Mechanism of A-Motility
The molecular characterization and localization of major motility proteins strongly argues that A-motility is propelled by a periodically assembled transenvelope complex that links a molecular motor in the bacterial inner membrane to the outer substrate (Figure 6.2b) . However, the exact mechanism by which the Agl–Glt complex powers motility remains to be determined. Ultimately, solving the mechanism requires answers to several outstanding questions: What is the mechanism of force transduction and how does the complex transmit forces through the rigid cell wall? What is the mechanism of directionality and is there a track for the motility complex? How does the complex connect with the surface?
Evidence for the Existence of a Track
The Agl–Glt machinery is thought to act as a stator that propels the cell as it threads along a rotor connected to rigid scaffolds in the cell. The bacterial MreB-actin cytoskeleton was initially proposed to constitute the rigid track because MreB depolymerization with the MreB-specific inhibitor A22 rapidly and reversibly blocks A-motility (Mauriello et al. 2010) . Additionally, polystyrene beads and AglR motions are also blocked by the action of A22 (Nan et al. 2013; Sun et al. 2011) . However, even though MreB may be essential for motility, it may not function as a motility rotor. First, the structure of the AglRQS motor suggests that it functions to generate a power stroke in the periplasmic space and not in the cytosolic compartment where MreB is localized. As mentioned above, AglRQS is a TolQR-like complex and this complex energizes envelope processes in the periplasmic space. Specifically, in the Tol-Pal system, the interaction between TolQR and TolA, the suspected energy transducer, likely allows dynamic contacts with the outer membrane through the Pal lipoprotein (Cascales et al. 2000) . Since GltG is a TolA homologue, the power stroke of the AglRQS likely occurs in the periplasm. Second, in bacteria, MreB is centrally linked to the synthesis of new peptidoglycan (PG; Domínguez-Escobar et al. 2011) . Recently, MreB has been shown to form short patch-like bundles rather than a continuous helix as initially thought (Domínguez-Escobar et al. 2011; Garner et al. 2011; van Teeffelen et al. 2011) . Therefore, it is unlikely that MreB could form a continuous track in Myxococcus cells. MreB may function to position the motility complex or allow its insertion in the PG or both.
Recent high-resolution tracking of single AglR particles suggested that they follow trajectories consistent with a helical path (Nan et al. 2013) . Consistent with this, deconvolution microscopy suggests that GltD-mCherry forms a closed loop structure in the bacterial cell envelope (Figure 6.3c; Nan et al. 2011) . However, deconvolution is prone to many artifacts and apparent helical structures may be interpreted with a grain of salt as shown by earlier interpretations of MreB helices. If the rigid track is a helix , the cell body should rotate with respect to its point of attachment to the substratum, which could be tested experimentally.
Contact with the Substrate—The Role of Slime
Although slime deposition by gliding Myxococcus cells was observed as early as in the 1940s, its exact function has remained largely mysterious (Beebe 1941) . Recently, Ducret et al. (2012) developed a new imaging method called surface enhanced ellipsometric contrast microscopy in wet condition (Wet-SEEC) to image slime deposition at high resolution. This analysis revealed that slime is deposited at constant rates underneath the cell body and that during motility, slime patches are specifically bound by the Agl–Glt gliding machinery (although the secretion of slime does not depend on the motility machinery, Figure 6.3b). These observations suggested that slime acts as a self-secreted substrate, favoring the connection between the outermost components of the motility machinery and the underlying surface. The exact composition of slime is unknown but it appears to be composed of major carbohydrate polymer. More work is needed to determine the exact slime composition and how it becomes bound by the motility machinery.
The Current A-Motility Model
During the past 5 years, the understanding of A-motility has made a tremendous leap forward and a new updated model can be proposed (Figure 6.4a). The motility machinery is composed by a stator formed by AglRQS, which is anchored in the bacterial inner membrane, and a rotor consisting of Glt proteins and/or the MreB cytoskeleton . Following their assembly at the leading cell pole, active motor AglRQS units become loaded with Glt proteins and the resulting complex then moves directionally along the helical track of the rotor toward the lagging cell pole. When the motility machinery reaches the face of the cell that is in direct contact with the substrate, it becomes immobilized locally (hence forming focal adhesions) by its interaction with slime , creating a drag force that propels the cell body forward. Persistent movements would result from an inherent cell asymmetry ensuring that the Agl motor moves along the helical track in two conformations: as an active motility machinery, when it is loaded with the Glt complex and moves from the leading cell pole to the lagging cell pole; or as an inactive machinery as it moves back in the opposite direction from the lagging pole to the leading pole. This idea is supported by the existence of pole-specific motility proteins that discriminate the leading and the lagging pole (Leonardy et al. 2010; Zhang et al. 2010) . At the leading cell pole, a Ras-like small G-protein MglA is essential for the polar assembly of the motility complex, while its negative regulator, MglB is required for the inactivation of the motility complex at the opposite pole (Leonardy et al. 2010; Zhang et al. 2010) .
The Myxococcus xanthus gliding motility model . a The red arrow represents the direction of movement. Motility motors loaded with the Glt complex (big dark red circles) or unloaded motors (small transparent red circles) translocate along an endless closed loop. Only the motors loaded with the Glt complex are proficient for movement. The machinery could afford the rigid PG by two different ways: b The motility complex may span the entire cell envelope and a PG-hydrolase may facilitate insertion of the complex through the PG. c Alternatively, the motility complex could deform the PG, creating surface depression and drag. Outer membrane proteins may, in this system, reinforce local contacts at the depressions
Given that the motility motor is located in the bacterial inner membrane, it must interact with the substrate beyond the rigid bacterial cell wall. How this connection occurs is undetermined. At the point of attachment, the motility complexes could span the entire cell envelope (inner membrane, PG, and outer membrane) and engage elastic interactions with slime and the substrate with specific outer membrane proteins (Figure 6.4b). This hypothesis is supported by the presence of gliding proteins in each envelope layer (Figure 6.2b; Luciano et al. 2011) , and by the observation that the slime is directly connected to the Agl–Glt machinery (Figure 6.3a, b; Ducret et al. 2012) . However, such attachment implies that the Agl–Glt complex forms a continuous envelope-spanning complex that pokes through the PG layer and must traverse it at all times. This difficulty would be solved if the motility machinery is associated with a PG-degrading activity, i.e., a hydrolase, to degrade the PG locally, allowing its insertion in the PG (Figure 6.4b). Such hydrolase activity is yet to be found. Alternatively, as discussed above, the interaction with MreB could facilitate the insertion of the gliding machinery in the PG.
In an alternative mechanism, the gliding machinery would not cross the cell wall but would distort it when it is loaded with the Glt complex (Nan et al. 2011) . This distortion would push against the outer membrane, literally creating bumps, and thus contact zones against the substratum (Figure 6.4c). Propulsion would then occur due to viscous coupling at the contact zones. Further experiments are needed to discriminate between these two possibilities, on one hand, total internal reflection fluorescence (TIRF) microscopy suggests the existence of periodic undulations in the bacterial envelope in contact with the substrate , which is consistent with the viscous coupling mechanism (Nan et al. 2011) ; and on the other hand, the Glt proteins comprise several critical outer membrane proteins that should only be important if the coupling is elastic (Luciano et al. 2011) .
6.2 Gliding Motility in the Bacteroidetes
Gliding motility is largely represented in the Bacteroidetes phylum, where it has been mostly studied in the Cytophagales and the Flavobacteriales . In this bacterial branch, gliding motility contributes to a number of cell behaviors and environmental adaptations (Jarrell and McBride 2008) . Recently, gliding motility has been shown to contribute to the formation of uniquely structured biofilms with iridescent properties in Cellulophaga spp. (Kientz et al. 2012) . At the molecular level, the mechanism of gliding motility has been mostly studied in Flavobacterium. Although many features of the gliding mechanism resemble gliding features in Myxococcus xanthus , there are also key differences. One fundamental difference is the speed of gliding in the µm/s range, which thus far exceeds the speed of Myxococcus gliding motility (µm/min), suggesting major differences in the motility engines. In this part of the chapter, we discuss recent progress in the study of Flavobacterium johnsoniae motility. Contrary to Myxococcus xanthus, the Flavobacterium motility engine has not been identified. But overall, the evidence suggests a propulsion mechanism involving the helical trafficking of outer-membrane adhesins and the involvement of a new type of secretion system .
6.2.1 Flavobacterium gliding involves a repertoire of outer membrane adhesins
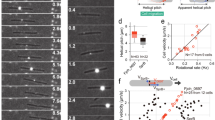
At the cell surface, Flavobacterium gliding motility involves two major adhesins, SprB and RemA (Figs. 6.5 and 6.6a). SprB, a huge protein (669 kDa), is probably the major adhesin and is required for gliding on agar and to some extent gliding on glass (Nelson et al. 2008) . The binding activity of RemA is only unmasked when sprB is deleted, suggesting that it plays partially redundant functions (Shrivastava et al. 2012) . SprB is a large repetitive cell-surface protein with an extensive beta-sheet structure. When observed by electron microscopy on whole cells, SprB is readily visible as thin filamentous spikes that extrude outwards from the cell surface (Figure 6.5a; Nakane et al. 2013) . How this conformation is linked to SprB function is unknown.
Helical motions of the SprB adhesin. a SprB forms cell-surface filaments. Negative staining of wild-type and sprB deletion strains (reprinted with permission from Nakane et al. 2013). b Polystyrene spheres coated with anti-SprB move rapidly along the cell surface (reprinted with permission from Nelson et al. 2008). c Location of SprB observed by TIRF microscopy. A cell translocating to the right was analyzed. The SprB signals were colored from red to blue at 0.05-s intervals for 1.25 s and integrated into one image (Lower). (reprinted with permission from Nakane et al. 2013)
Gliding mechanism of F. johnsoniae. a Proteins involved in F. johnsoniae gliding motility and protein secretion. SprB and RemA (orange) are thought to function as adhesins that are propelled along the cell surface by the some of the other proteins shown. GldA, GldF, and GldG (red) comprise an ATP-binding cassette transporter whose exact role in gliding is not known. GldI (yellow) is a peptidylprolyl isomerase involved in protein folding. Proteins in green (GldK, GldL, GldM, GldN, SprA, SprE, SprF, SprT) constitute the PorSS and are required for secretion of SprB and RemA and for motility. They also secrete the chitinase ChiA (white), which is not involved in motility. Proteins secreted by the PorSS have a predicted type-1 signal peptides and are predicted to be exported across the cytoplasmic membrane by the Sec system before being secreted across the outer membrane by the PorSS. Proteins in blue (GldB, GldD, GldH, and GldJ) are also required for gliding. Black lines indicate lipid tails on lipoproteins. b Flavobacterium gliding is thought to be powered by motors composed of Gld proteins in the cell envelope that propel adhesins, such as SprB, along the cell surface. Adhesin SprB moves along the left-handed helical loop and has two different states: SprB moving toward the front of the cell and SprB moving toward the rear of the cell. In a translocating cell, SprB moving toward the rear of the cell adheres to the surface, generating left-handed rotation and right-directed translocation of the cell
RemA is likely a polysaccharide-binding protein because it contains a lectin-type domain (Shrivastava et al. 2012) . Consistent with this, liquid-grown cells clump in liquid cultures while an remA mutant does not. This suggests that RemA binds to a self-produced polysaccharide, a potential equivalent to the Myxococcus slime. Interestingly, the same genetic screen that led to the identification of RemA also identified three additional genes, remC, wza, and wzc (Shrivastava et al. 2012) . remC gene encodes a putative glycosyltransferase and wza and wzc encode the octameric secretion pore and the inner membrane parts of a so-called outer membrane auxiliary (OMA or Wza), a capsular polysaccharide secretion system. Since mutations in all three genes failed to form large cell aggregates, it is tempting to suggest that the substrate of this Wza system interacts with RemA. Additional RemA-like adhesins might participate to the motility process because many proteins of this type are encoded in the Flavobacterium genome.
6.2.2 The Gliding Motility Mechanism Involves Helical Trafficking of the Surface Adhesins
The evidence that the motility mechanism is linked to the directed motion of protein complexes at the cell surface was first provided by Lapidus and Berg, who in the early 1980s observed that particles are propelled at velocities matching the gliding speed when they are bound to the Cytophaga cell surface (Lapidus and Berg 1982) . However, at that time, video microscopy techniques did not allow high-resolution tracking of the particles and the complex trajectories could not be computed into a coherent motility model. The recent identification of proteins such as RemA and SprB provided a new opportunity to track the surface dynamics of the motility components. Indeed, both RemA and SprB move directionally at the cell surface with trajectories that resemble the particle movements described by Lapidus and Berg (Figure 6.5b; Nakane et al. 2013; Shrivastava et al. 2012) .
Recently, the dynamics of SprB were resolved at high resolution by TIRF microscopy of moving Flavobacterium cells (Nakane et al. 2013) . It was thus found that SprB moves directionally along the length of the cell in a left-handed helical manner (Figure 6.5c). Two types of dynamic behaviors were observed: when SprB subunits moved from the leading to the lagging cell pole, they mostly formed fixed sites reminiscent of the Myxococcus focal adhesions. When SprB subunits reached the back of the cell, they were observed to move back to the leading cell pole along the same helical structure at a speed that matched the cell velocity. Similar to Myxococcus xanthus , gliding motility may be propelled by a helical machinery where active motility complexes travel directionally towards the lagging cell pole (Figure 6.6b). In the active state, corresponding to SprB moving in the anteroposterior direction, SprB presumably attaches to the substratum, immobilizing the cell body and thus propelling the cell forward. Upon reaching the rear of the cell, substrate-attached SprB adhesins are released from the substratum and recycled back to the front of the cell. How this inactivation occurs is unknown. SprB is only seen moving relative to the cell body in the posteroanterior direction. This implies that SprB adhesins have been modified at the cell rear to prevent their attachment when moving in the opposite direction (Figure 6.6b). Fundamentally, this propulsion mechanism is very similar to the propulsion mechanism of Myxococcus; however, the two systems are also very different, largely because the motility machineries at work are distinct.
6.2.3 The Flavobacterium Gliding Motility Genes and the Still Elusive Gliding Machinery
Over the years, the search of motility mutants has uncovered a total of 17 genes that appear to form the core of the motility complex (named gldA, B, D, F, G, H, I, J, K, L, M, N, sprA, B, E, T, and remA, Figure 6.6a; McBride and Zhu 2013) . Many of these genes are only found in genomes of the Bacteroidetes phylum, suggesting that Bacteroidetes gliding employs a unique machinery (McBride and Zhu 2013) . Although it appears that the structural core components of the motility machinery are likely encoded by some of the gld genes, the structure of the actual machinery remains unknown mostly because the motility motor cannot be predicted from annotations of the gld genes and a subset of the gld and spr genes encode a new secretion system, the PorSS or type-IX secretion system (T9SS). This PorSS may have an accessory function in the secretion of key motility proteins, for example the terminal adhesins SprB and RemA (Sato et al. 2010; Shrivastava et al. 2013) .
6.2.4 Gliding Motility Requires a New Secretion System and an Unknown Motor
In 2010, studies in Porphyromonas gingivalis, a periodontal pathogen from the Bacteroidetes subgroup, revealed that key virulence factors, such as the gingipains, are secreted by a novel secretion system (PorSS or T9SS) that is unique to the Bacteroidetes phylum (McBride and Zhu 2013; Sato et al. 2010) . Remarkably, several of the newly identified Porphyromonas PorSS genes were paralogs to a subset of the gld and spr genes (gldK, L, M, N and sprA, E, T; Figure 6.6a). In Flavobacterium, these particular genes are required for the secretion of SprB and RemA, two essential motility outer membrane adhesins, suggesting that they also encode a PorSS-type secretion apparatus (Shrivastava et al. 2013) . This finding potentially explains the long known coupling between motility and the ability of Flavobacterium to degrade chitin, because a critical chitinase is also secreted by the T9SS (Kharade and McBride 2014) . Remarkably, the substrates of the T9SS contain a specific C-terminal domain (CTD) that seems important for their targeting to the secretion apparatus.
It is not presently clear whether the Bacteroidetes gliding machinery evolved by modular expansion of the T9SS or whether the T9SS and gliding machinery operate independently, the T9SS only being essential for gliding because it secretes the terminal motility adhesins. In bacteria, motility and secretion systems are often evolutionarily connected, conspicuously, the flagellum and type-III secretion system (T3SS; Abby and Rocha 2012) and the T4P and type-II secretion system (T2SS; Pelicic 2008) . In Myxococcus, the Agl–Glt system probably evolved through the specialization of a general class of surface transporters (Luciano et al. 2011) , themselves evolved from a simpler core apparatus of unknown function, possibly a protein secretion system. By analogy, the Bacteroidetes gliding machinery may well be modular and contain a T9SS for its assemblage and function.
To answer this question, it will be essential to characterize the protein system that energizes motility. As discussed above, flagellar stator homologues power Myxococcus gliding motility . In Flavobacterium, several studies also indicate that the PMF could be the energy source for gliding motility (Dzink-Fox et al. 1997; Nakane et al. 2013) . Specifically, Dzink-Fox et al. showed that acetate, a protonophore known to dissipate the PMF, inhibits cell movements (Dzink-Fox et al. 1997). This effect is unlikely the indirect consequence of a block in T9SS secretion because the surface movements of SprB is blocked rapidly and reversibly by CCCP, which also dissipates the PMF across the cytoplasmic membrane (Nakane et al. 2013) . Thus, similar to Myxococcus, proton-conducting channels of the inner membrane may energize motility. However, amongst the known Gld proteins, none have canonical features of flagellar Mot or Myxococcus Agl proteins. The motor genes may still need to be identified but it is also possible that the PMF acts at another level of the motility process and alternatively, ATP could fuel the motility engine. GldF and GldG were initial motor candidates because they are predicted components of an ATP-binding cassette transporter. However, although GldF and GldG are required for F. johnsoniae gliding, they are not present in all gliding Bacteroidetes phylum, suggesting that they are not core components of the gliding motility machinery (McBride and Zhu 2013) .
In the future, it will be critical to identify the motility motor, which presumably localizes to the bacterial inner membrane (Nakane et al. 2013) . Whatever the exact identity of the motor and its source of energy, it must span the bacterial inner membrane and transduce its activity beyond the periplasmic space to the outer membrane adhesins. In Flavobacterium, interactions with the surface are mediated by outer membrane adhesins, which clearly implies that the molecular motor must interact with the outer membrane through the PG. How this occurs is unknown. The evidence for the existence of a helical track is strong but the identity of that track is unknown.
6.3 General Conclusions
In recent years, tremendous progress has been made in the understanding of bacterial gliding motility , this long mysterious process occurring in absence of visible extracellular appendages. Studies from Myxococcus and Flavobacterium provide key and complementary pictures to understand this complex cellular behavior. Work in Myxococcus has tremendously improved our understanding of the intracellular protein dynamics that underlie the gliding process. Clearly, the local and transient immobilization of inner membrane motors and force transduction through the cell envelope is critical for cell motion. However, in this system, the exact propulsion mechanism remains unclear. Circumstantial evidence, such as the potential helical arrangement of GltD and possible rotational motions of motor subunits, suggest the existence of a helical rotor. In Flavobacterium, the helical motion of motility adhesins is strongly supported, further suggesting that bacterial gliding motility is linked to rotational helical tracks in the cell envelope. While this exciting parallel suggests a common mechanism, it should be considered with cautious because the gliding motility machineries are not identical and there are tremendous differences in the gliding motility speeds between the two organisms. Further understanding thus awaits that the gliding machinery and motor be identified in Flavobacterium. Similarly, the outer membrane dynamics of the Myxococcus Agl–Glt machinery will have to be characterized to confirm the existence of the helical rotor.
Beyond the understanding of the exact motility mechanism, studies of bacterial gliding motility will likely deeply impact our understanding of protein movements and dynamics of the bacterial cell envelope . In this direction, studies of Myxococcus motility have already shown that the motility machinery is related to a new class of transport systems that contribute to the surface anchoring of capsular-type exopolysaccharides (Wartel et al. 2013) .
References
Abby SS, Rocha EPC (2012) The non-flagellar type III secretion system evolved from the bacterial flagellum and diversified into host-cell adapted systems. PLoS Genet 8:e1002983
Beebe JM (1941) The morphology and cytology of Myxococcus xanthus. N Sp J Bacteriol 42:193–223
Burchard RP (1981) Gliding motility of prokaryotes: ultrastructure, physiology, and genetics. Annu Rev Microbiol 35:497–529
Cascales E, Gavioli M, Sturgis JN, Lloubès R (2000) Proton motive force drives the interaction of the inner membrane TolA and outer membrane pal proteins in Escherichia coli. Mol Microbiol 38:904–915
Dhahri S, Ramonda M, Marlière C (2013) In-situ determination of the mechanical properties of gliding or non-motile bacteria by atomic force microscopy under physiological conditions without immobilization. PLoS ONE 8:e61663
Domínguez-Escobar J, Chastanet A, Crevenna AH, Fromion V, Wedlich-Söldner R, Carballido-López R (2011) Processive movement of MreB-associated cell wall biosynthetic complexes in bacteria. Science 333:225–228
Ducret A, Valignat M-P, Mouhamar F, Mignot T, Theodoly O (2012) Wet-surface-enhanced ellipsometric contrast microscopy identifies slime as a major adhesion factor during bacterial surface motility. Proc Natl Acad Sci U S A 109:10036–10041
Dzink-Fox JL, Leadbetter ER, Godchaux W 3rd (1997) Acetate acts as a protonophore and differentially affects bead movement and cell migration of the gliding bacterium Cytophaga johnsonae (Flavobacterium johnsoniae). Microbiol Read Engl 143(Pt 12):3693–3701
Garner EC, Bernard R, Wang W, Zhuang X, Rudner DZ, Mitchison T (2011) Coupled, circumferential motions of the cell wall synthesis machinery and MreB filaments in B. subtilis. Science
Hodgkin J, Kaiser D (1979) Genetics of gliding motility in Myxococcus-Xanthus (Myxobacterales)—2 gene systems control movement. Mol Gen Genet 171:177–191
Hoiczyk E, Baumeister W (1998) The junctional pore complex, a prokaryotic secretion organelle, is the molecular motor underlying gliding motility in cyanobacteria. Curr Biol 8:1161–1168
Jarrell KF, McBride MJ (2008) The surprisingly diverse ways that prokaryotes move. Nat Rev Microbiol 6:466–476
Kaiser D (2003) Coupling cell movement to multicellular development in myxobacteria. Nat Rev Microbiol 1:45–54
Kharade SS, McBride MJ (2014) Flavobacterium johnsoniae chitinase ChiA is required for chitin utilization and is secreted by the type IX secretion system. J Bacteriol 196:961–970
Kientz B, Ducret A, Luke S, Vukusic P, Mignot T, Rosenfeld E (2012) Glitter-like iridescence within the bacteroidetes especially Cellulophaga spp.: optical properties and correlation with gliding motility. PLoS ONE 7:e52900
Lapidus IR, Berg HC (1982) Gliding motility of Cytophaga sp. strain U67. J Bacteriol 151:384–398
Leonardy S, Miertzschke M, Bulyha I, Sperling E, Wittinghofer A, Sogaard-Andersen L (2010) Regulation of dynamic polarity switching in bacteria by a Ras-like G-protein and its cognate GAP. Embo J 29:2276–2289
Li Y, Sun H, Ma X, Lu A, Lux R, Zusman D, Shi W (2003) Extracellular polysaccharides mediate pilus retraction during social motility of Myxococcus xanthus. Proc Natl Acad Sci U S A 100:5443–5448
Luciano J, Agrebi R, Le Gall AV, Wartel M, Fiegna F, Ducret A, Brochier-Armanet C, Mignot T (2011) Emergence and modular evolution of a novel motility machinery in bacteria. PLoS Genet 7:e1002268
Mauriello EM, Nan B, Zusman DR (2009) AglZ regulates adventurous (A-) motility in Myxococcus xanthus through its interaction with the cytoplasmic receptor, FrzCD. Mol Microbiol 72:964–977
Mauriello EMF, Mouhamar F, Nan B, Ducret A, Dai D, Zusman DR, Mignot T (2010) Bacterial motility complexes require the actin-like protein, MreB and the Ras homologue, MglA. EMBO J 29:315–326
McBride MJ, Zhu Y (2013) Gliding motility and Por secretion system genes are widespread among members of the phylum bacteroidetes. J Bacteriol 195:270–278
Mignot T (2007) The elusive engine in Myxococcus xanthus gliding motility. Cell Mol Life Sci 64:2733–2745
Mignot T, Shaevitz JW, Hartzell PL, Zusman DR (2007) Evidence that focal adhesion complexes power bacterial gliding motility. Science 315:853–856
Nakane D, Sato K, Wada H, McBride MJ, Nakayama K (2013) Helical flow of surface protein required for bacterial gliding motility. Proc Natl Acad Sci U S A 110:11145–11150
Nan B, Zusman DR (2011) Uncovering the mystery of gliding motility in the myxobacteria. Annu Rev Genet 45:21–39
Nan B, Mauriello EM, Sun IH, Wong A, Zusman DR (2010) A multi-protein complex from Myxococcus xanthus required for bacterial gliding motility. Mol Microbiol 76:1539–1554
Nan B, Chen J, Neu JC, Berry RM, Oster G, Zusman DR (2011) Myxobacteria gliding motility requires cytoskeleton rotation powered by proton motive force. Proc Natl Acad Sci U S A 108:2498–2503
Nan B, Bandaria JN, Moghtaderi A, Sun I-H, Yildiz A, Zusman DR (2013) Flagella stator homologs function as motors for myxobacterial gliding motility by moving in helical trajectories. Proc Natl Acad Sci U S A 110:E1508–E1513
Nelson SS, Bollampalli S, McBride MJ (2008) SprB is a cell surface component of the Flavobacterium johnsoniae gliding motility machinery. J Bacteriol 190:2851–2857
Pelicic V (2008) Type IV pili: e pluribus unum? Mol Microbiol 68:827–837
Sato K, Naito M, Yukitake H, Hirakawa H, Shoji M, McBride MJ, Rhodes RG, Nakayama K (2010) A protein secretion system linked to bacteroidete gliding motility and pathogenesis. Proc Natl Acad Sci U S A 107:276–281
Shrivastava A, Rhodes RG, Pochiraju S, Nakane D, McBride MJ (2012) Flavobacterium johnsoniae RemA is a mobile cell surface lectin involved in gliding. J Bacteriol 194:3678–3688
Shrivastava A, Johnston JJ, van Baaren JM, McBride MJ (2013) Flavobacterium johnsoniae GldK, GldL, GldM, and SprA are required for secretion of the cell surface gliding motility adhesins SprB and RemA. J Bacteriol 195:3201–3212
Skerker JM, Berg HC (2001) Direct observation of extension and retraction of type IV pili. Proc Natl Acad Sci U S A 98:6901–6904
Sun M, Wartel M, Cascales E, Shaevitz JW, Mignot T (2011) Motor-driven intracellular transport powers bacterial gliding motility. Proc Natl Acad Sci U S A 108:7559–7564
Van Teeffelen S, Wang S, Furchtgott L, Huang KC, Wingreen NS, Shaevitz JW, Gitai Z (2011) The bacterial actin MreB rotates, and rotation depends on cell-wall assembly. Proc Natl Acad Sci U S A 108:15822–15827
Wartel M, Ducret A, Thutupalli S, Czerwinski F, Le Gall A-V, Mauriello EMF, Bergam P, Brun YV, Shaevitz J, Mignot T (2013) A versatile class of cell surface directional motors gives rise to gliding motility and sporulation in Myxococcus xanthus. PLoS Biol 11:e1001728
Youderian P, Burke N, White DJ, Hartzell PL (2003) Identification of genes required for adventurous gliding motility in Myxococcus xanthus with the transposable element mariner. Mol Microbiol 49:555–570
Yu R, Kaiser D (2007) Gliding motility and polarized slime secretion. Mol Microbiol 63:454–467
Zhang Y, Franco M, Ducret A, Mignot T (2010) A bacterial Ras-like small GTP-binding protein and its cognate GAP establish a dynamic spatial polarity axis to control directed motility. PLoS Biol 8:e1000430.
Zhang Y, Ducret A, Shaevitz J, Mignot T (2012) From individual cell motility to collective behaviors: insights from a prokaryote, Myxococcus xanthus. FEMS Microbiol Rev 36:149–164
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Wartel, M., Mignot, T. (2014). The Mechanism of Bacterial Gliding Motility: Insights from Molecular and Cellular Studies in the Myxobacteria and Bacteroidetes. In: Barton, L., Bazylinski, D., Xu, H. (eds) Nanomicrobiology. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1667-2_6
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1667-2_6
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1666-5
Online ISBN: 978-1-4939-1667-2
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)