Abstract
The dominant driver of malignant behavior for most prostate cancers is the androgen receptor (AR). AR signaling may act at the genesis of the disease to trigger the appearance of rearrangements between AR-regulated genes, such as TMPRSS2, and transcription factor genes, such as ERG, and in turn, the products of such fusion genes, now regulated by AR, function to prevent terminal differentiation and to promote invasive tumor growth and metastatic dissemination. Interference with AR function has served as the principal treatment for advanced prostate cancer for more than seven decades. However, despite the well-known benefits of androgen deprivation therapy, disease progression to castration-resistant prostate cancer (CRPC) has proven inevitable. CRPC appears to evolve via two major mechanisms: one with maintained AR signaling (AR “addiction”) and the other without (escape from AR “addiction”). In this chapter, the molecular features of AR “addicted” and non-AR “addicted” cancers will be reviewed.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- prostate cancer
- androgen receptor (AR)
- castration-resistant prostate cancer (CRPC)
- enzalutamide
- abiraterone
- neuroendocrine prostate cancer (NEPC)
Almost all prostate cancers acquire an addiction to androgenic hormones during disease development. In the normal prostate, the testicular androgen testosterone (T) is converted to the more potent androgen dihydrotestosterone (DHT) to promote gland secretory function. Androgen regulation of prostatic differentiation is then accomplished by DHT binding to an intracellular androgen receptor (AR), which triggers a cascade of events culminating in translocation of the receptor into the cell nucleus and trans-activation of key differentiation genes, including KLK3 (encoding prostate-specific antigen [PSA]) and TMPRSS2 [1–3]. Prostate cancer cells become addicted to this signaling pathway by co-opting the AR to drive malignant behavior(s). In doing so, the cells maintain a caricature of a secretory cell phenotype, producing PSA and secreting it into the bloodstream rather than the ejaculate. At the same time, the cells are able to use AR to escape the limits of terminal differentiation. For this reason, the lowering of circulating androgen levels by treatment with bilateral orchiectomy, estrogens, or gonadotrophin releasing hormone (GnRH) analogs has long been used to treat advanced prostate cancers [4]. In nearly all cases, this maneuver results in a fall in serum PSA levels and an improvement in symptoms attributable to prostate cancer. Unfortunately for nearly all men, inexorable progression of disease to “castration-resistant prostate cancer (CRPC)” ensues. Emerging evidence suggests that CRPC comprises a heterogeneous collection of cancers, some cases with an ongoing addiction to AR signaling, potentially treatable with new drugs like abiraterone and enzalutamide, and other cases that have become AR-independent [5]. In this chapter, the molecular mechanisms responsible for these CRPC phenotypes will be reviewed.
Gene Fusions and Prostate Cancer Dependence on AR
Somatic chromosomal translocations and deletions creating gene fusions appear most likely responsible for subverting AR-dependent terminal differentiation in prostate cancer cells, permitting AR signaling to foment inappropriate cell growth and survival, invasiveness, and metastasis. The most common such genome alteration, generating a fusion between TMPRSS2, an AR-regulated prostate differentiation gene, and ERG, an ETS family transcription factor gene, has been found in up to half of prostate cancer cases [6–8]. The resultant dysregulated ERG expression directly endows prostate cancer cells with malignant properties such as invasiveness [9, 10]. In addition, ERG also indirectly undermines AR-dependent differentiation by interacting with AR at selected sites in the genome, interfering with AR trans-activation and allowing trans-repression via activation of EZH2, the polycomb repressor component endowed with H3K27 methyltransferase activity [11]. This action of ERG does not appear to reflect a general antagonism of AR signaling per se: in the setting of PTEN loss, which otherwise tends to result in a general dampening of AR target gene expression in prostate cells, ERG augments the general output of the AR signaling pathway [12, 13].
Forced ETS transcription factor expression in mouse prostate cells carrying disrupted Pten genes leads to highly penetrant invasive adenocarcinoma [13]. In this setting, the ETS factors collaborate with AR to increase the expression of many genes regulating invasion/migration, angiogenesis, and cell death [13]. Thus, fusion genes creating AR-regulated ETS factors perturb AR signaling in prostate cancer cells in a nuanced manner, preventing terminal differentiation while permitting inappropriate activation of genes associated with malignancy. In this way, prostate cancer cells are addicted to the AR, which becomes needed both for ETS fusion gene expression and for the collaborative regulation of other malignancy genes. Not surprisingly, this addiction may be difficult to shake, as the cooperation between AR and ETS factors appears to confer robust tolerance to the deleterious consequences of additional somatic gene defects, such as inappropriate activation of PI3K-signaling accompanying PTEN loss, that might be acquired during prostate cancer progression, even to CRPC.
ERG is not normally expressed in prostatic epithelial cells; its appearance in such cells almost always reflects a somatic gene accident. The translocations and deletions allowing the AR-stimulated expression of ERG (and other cancer genes) in prostate cancer cells bring the androgen response element (ARE)-containing DNA sequences in the promoter and enhancer regions of TMPRSS2 (and other AR-regulated genes) into continuity with ERG coding sequences [6, 8]. Remarkably, such chromosomal rearrangements appear to be triggered by AR itself. To initiate transcription of target genes, ligand-bound AR builds a transcription complex by engaging co-activators and by altering chromatin conformation. As part of this process, AR binds TOP2B, a DNA topoisomerase capable of double strand passage, to prevent tangling during DNA template looping and migration to transcription “factory” sites in the cell nucleus [14]. TOP2B function is vital to the initiation of transcription at AR gene targets, as knockdown of TOP2B expression or inhibition of TOP2B enzymatic activity prevents AR-dependent gene expression [14]. When it recruits TOP2B to the transcriptional regulatory region of genes like TMPRSS2, AR tends to stimulate TOP2B-mediated DNA double strand breaks that can be substrates for illegitimate recombination upon repair by the non-homologous end joining (NHEJ) pathway [14, 15]. TOP2-triggered DNA strand breaks have been implicated in the generation of gene fusions involving the MLL gene in treatment-associated acute myeloid leukemia (t-AML; see [16]). In this setting, enzyme dysfunction was likely induced as a consequence of exposure to TOP2-targeted anti-neoplastic drugs. However, the chromosomal rearrangements in prostate cancer cells may arise as a result of a more intrinsically error prone process. In model studies using prostate cancer cells, AR-triggered TOP2B activation can promote recombination at or near ARE sequences in AR target genes and create TMPRSS2–ERG fusion transcripts [14].
The contributions of AR and androgen signaling to the malignant phenotype of prostate cancer cells cannot be overstated. AR acts to induce translocations and deletions engendering gene fusions [14]. The dysregulated products of fusion genes then undermine AR-associated terminal differentiation, collaborate with AR to activate tumorigenic pathways, and prevent cell death associated with oncogenic stress, and reinforce AR-transcriptional output during prostate cancer progression [9–13]. In this way, AR action both promotes genetic instability and malignant behavior, and fusion genes facilitate co-opting of AR signaling to maintain the cancer phenotype. This type of mechanism predicts that interference with AR function should be deleterious to prostate cancer cells, underscoring the well-recognized benefit of androgen deprivation to prostate cancer treatment, and that progression to CRPC could conceivably proceed via two different routes: (1) by maintaining AR signaling in some manner, or (2) by through the development of a molecular escape mechanism from AR addiction (Table 3.1).
The Molecular Biology of AR Function
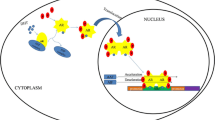
AR is a ligand-dependent transcription factor encoded by a single gene with 8 exons located at Xq11-12. The receptor is a member of the nuclear receptor superfamily group that also contains the glucocorticoid, mineralocorticoid, and progesterone receptors 3 [17]. The physiologic AR protein shows marked inter-individual differences in size as a result of variable polyglutamine and polyglycine repeats. These differences may affect receptor function, with both increased transcriptional trans-activation and increased prostate cancer risk seen in association with AR containing shorter polyglutamine repeats [18, 19]. AR structure can be considered in terms of mapped functional domains, including a ligand binding domain (LBD) ensuring selective activation by androgenic hormones, a hinge domain, a DNA binding domain (DBD) permitting binding selective binding to ARE sequences, and an N-terminal domain; critical regions for transcription trans-activation (activation function or AF regions) are located both in the LBD and in the N-terminal region [1]. In the absence of androgens, the receptor is sequestered in the cytoplasm via an interaction with chaperone proteins. Hormone binding triggers a cascade of events starting with a change in AR conformation which results in liberation from chaperones, dimerization, and ingress into the cell nucleus [1].
The arrival of the ligand-bound, activated, AR in the cell nucleus attracts a myriad of transcriptional co-regulators, including histone acetyltransferases, histone demethylases, SWI/SNF proteins, poly(ADP-ribose) polymerase (PARP), and as mentioned, TOP2B, along with as many as 200 or more other proteins represented in several complexes [1, 14, 20, 21]. These complexes then act to modify chromatin proteins so as to sculpt an active chromatin conformation capable of loading RNA polymerase II at target genes. The activated AR is competent to directly engage co-regulatory proteins containing an FxxLF amino acid motif upon ligand binding as a result of the movement of helix 12 in the LBD to create a hydrophobic pocket (AF-2 [22]). The AR itself has an FQNLF amino acid sequence within its N-terminal domain, and androgen binding to the receptor can trigger a dimeric N-terminal to C-terminal conformation by virtue of interactions of FQNLF with the unveiled hydrophobic region in the LBD [20]. AR complexes bind genomic DNA not only at ARE sequences located in proximal promoters of genes but also at enhancer elements located far upstream, in introns, and in 3’ untranslated regions [23]. Once in the cell nucleus, activated AR can also modulate a number of genome functions, including facilitating the activation or repression of other genes and promoting licensing of DNA replication origins [24]. Of note, ligand-bound, activated, AR remaining in the cytoplasm has been reported to interact with kinases such as SRC to initiate additional signaling programs [25]. The transcription output attributable to activated AR in normal prostate cells includes differentiation genes such as KLK2, KLK3, and TMPRSS2, while in CRPC cells, AR also tends to promote expression of cell-cycle genes such as CDC20, UBE2C, CDK1, and ANAPC10 [23].
Molecular Mechanisms of Maintained AR Addiction in Many Cases of CRPC
Despite frequent initial beneficial treatment responses to androgen deprivation therapies that lower circulating testosterone levels to <50 ng/mL, CRPC tends eventually to emerge and progress to ultimately threaten life. Intriguingly, CRPC is most often heralded by progressive rises in serum PSA, a biomarker requiring activated AR function in prostate cancer cells [26]. This implies that there is ongoing AR signaling in these cancers, hinting they likely have remained addicted to AR [5]. How does this addiction persist? One mechanism involves ongoing production of androgens, either by adrenals or by the cancer itself, which persists despite ablation of testicular androgen biosynthesis [27, 28]. At cancer sites, T and DHT can be produced at levels sufficient to activate AR either by conversion of adrenal androgen precursors or by new synthesis using CYP17 [27, 28]. This process can be antagonized by ketoconazole, an antifungal drug capable of inhibiting CYP17 when administered at high doses, and by abiraterone acetate, a pregnenolone analog now approved for treatment of CRPC based on survival prolongation in randomized clinical trials [29]. A second CYP17 inhibitor, orteronel (TAK-700), is in advanced clinical development [30].
Another AR-addicted CRPC phenotype can be attributed to new AR mutations, not present at disease presentation, which encode receptors with altered ligand specificity. These cases tend to arise in the setting of treatment with “first-generation” anti-androgens, such as flutamide and bicalutamide, and may be responsible for an “anti-androgen withdrawal” syndrome, where men with disease progression despite the combination of androgen deprivation and anti-androgen administration show improvement upon cessation of anti-androgen treatment but maintenance of androgen deprivation [6, 31, 32]. Such AR mutations do not arise commonly in the setting of androgen deprivation alone. Even in the absence of AR mutations AR-addicted CPRC cases may arise as a consequence of ongoing AR activation stimulated by growth factor signaling pathways capable of creating post-translational modifications of AR or its co-activators [1, 5, 26].
In a study of CRPC arising among several different human prostate cancer xenografts propagated in immunodeficient mice, the most consistent molecular finding was that the abundance of AR was increased, leading to augmented transcriptional trans-activation at lower androgenic hormone levels and in response to a more promiscuous collection of ligands [33]. Amplification of AR has been reported in as many as 80 % of CPRC cancers, with some 30 % exhibiting marked gene amplification, which may account for some instances of AR over-expression [34, 35]. This high AR expression phenotype was exploited in the discovery of the “second-generation” anti-androgens enzalutamide and ARN-509, drugs that appear capable of interfering with AR function despite high-level AR expression in prostate cancer cells that otherwise do not respond to “first-generation” anti-androgen treatment [36, 37]. When compared to the “first-generation” anti-androgens, these new agents exhibit less mixed agonist and antagonist activity when docking to the LBD, triggering a different location of helix 12. The drugs stop AR activation almost entirely, before receptor trafficking to the cell nucleus and binding to ARE sequences [36, 37]. Thus far, enzalutamide has gained approval for CRPC treatment based on randomized trial data showing a survival benefit and ARN-509 trials are ongoing [38].
Predictably, the growing use of enzalutamide for CRPC has fostered the emergence of cancers resistant to “second-generation” anti-androgens. In many such treatment-resistant cases, the AR signaling addiction appears maintained. Model studies of the acquisition of “second-generation” anti-androgen resistance using LNCaP prostate cancer cells have revealed a new mutant AR with an F876L amino acid change in the receptor LBD [39–41]. Enzalutamide was able to bind the F876L-AR with 48-fold greater affinity that wild-type AR; ARN-509 also bound F876L-AR [39]. When the F876L-AR was introduced into prostate cancer cells, enzalutamide and ARN-509 acted as agonists rather than as antagonists, driving AR target gene expression and stimulating prostate cancer cell growth in vitro and in vivo [39, 40]. The F876L-AR mutation has been detected in men with progressive CPRC despite “second-generation” anti-androgen treatment. When plasma DNA from a phase 1 clinical trial of ARN-509 for CPRC was assayed for AR mutations, 3 of 18 men with progressive serum PSA increases despite treatment were found to have a C to A missense change at AR nucleotide 2628 encoding the F876L-AR [39]. Already, drug discovery efforts are underway for “next-generation” anti-androgens which can inhibit signaling by F876L-AR, raising the possibility that as long as CRPC remains addicted to AR with an intact LBD, small molecule antagonist drug therapy may be feasible [41].
A final phenotype of maintained AR addiction in CRPC may be mediated by AR splice variants that encode receptors without LBDs that can nonetheless act to promote target gene transcription [42–46]. The variant AR transcripts contain deleted or cryptically inserted exons. Some such transcripts may be generated by defective AR genes arising in association with AR amplification or other AR rearrangements [47]. However, the majority of the variant AR transcripts likely arise a result of some sort of perturbation in transcription initiation and elongation rates that accompanies androgen deprivation [48]. The resultant receptors contain truncated C-terminus, with an intact N-terminal domain and DBD, but not a functioning LBD. Usually, in prostate cancer cells, the level of variant AR mRNA tends to be far less than that of full-length AR mRNA. However, this may underestimate the expression level of the truncated AR forms encoded by variant AR transcripts, which may be as high as 30 % or more of AR protein [49]. Forced expression of one such truncated receptor, AR-V7 which lacks an LBD and contains 16 amino acids from a cryptic exon, triggered expression cell-cycle regulatory genes in prostate cancer cells, whether or not androgens were present [50]. This AR variant, which has been detected in men with CRPC, can drive the growth of prostate cancer cells in the absence of androgenic hormones. Also, while both full-length AR and AR-V7 tend to promote expression of prostate differentiation genes like KLK3 and TMPRSS2, there are tantalizing differences, as yet unexplained, in the patterns of genes induced by each receptor [50]. Several of the AR variants so far detected also appear to be expressed in CPRC cells and to possess the propensity to propagate ligand-independent signals, while others may not act in this manner [20]. Nonetheless, data are accumulating to suggest that AR splice variants may contribute both to abiraterone resistance and to enzalutamide resistance in CRPC [51].
CRPC Abandonment of AR Addiction
Progression to CRPC, in part driven by highly unstable genomes and epigenetic regulation and in part driven by therapeutic pressure, can result in a phenotype/genotype that is entirely independent of AR signaling, a tumor state that has recently been operationally termed Androgen Receptor Pathway-Independent Prostate Cancer (APIPC) [52]. One type of APIPC can be characterized by features such as loss of PSA production, lytic bone metastases, hypercalcemia, or widely disseminated visceral metastases that are otherwise uncommon complications of systemically advanced prostate cancer. This prostate cancer cell phenotype, often referred to as neuroendocrine prostate cancer (NEPC), does not manifest AR expression or AR signaling, representing an escape from AR addiction [53]. Molecular “archeology” studies of NEPC strongly suggest that most such cases evolved from AR-addicted prostate cancers, as many have been found to contain TMPRSS2–ERG rearrangements [54]. Despite the presence of the rearrangements, the absence of AR signaling prevents TMPRSS2–ERG fusion mRNA or ERG protein expression. AR-independent NEPC cases appear instead to contain other genome alterations, such as amplification of N-MYC and AURKA [55]. For this reason, the NEPC variant of CRPC does not respond to treatments targeted at the AR signaling axis. Instead, the use of cytotoxic chemotherapy, usually with platinum compounds, is often attempted [56]. Ongoing clinical trials are assessing whether AURKA inhibitors might provide benefit to men with NEPC and AURKA amplification with AURKA over-expression. Of concern, though this AR-independent NEPC variant of CRPC was once thought to be rare, the frequency with which it appears may be increasing with the introduction of androgen biosynthesis inhibitors and “second-generation” anti-androgens [57]. CRPC progression to NEPC is clearly dangerous, autopsy studies of life-threatening CRPC have hinted that as many as 25 % or more of men who die with prostate cancer show signs of NEPC [58].
Recent evidence suggests prostate cancers transitioning from an AR-driven to an APIPC state are not obligated to progress via a neuroendocrine phenotype, but may be dependent on alternative survival pathways. For example, as many as 80 % or more of CRPC cases show increased FGF8 expression, and cases with amplification of FGFR2 have been identified [59, 60]. As men live longer with CRPC by responding to the new androgen biosynthesis inhibitors and better anti-androgens, more subtypes of APIPC are expected to emerge [61].
The Molecular Pathogenesis of Lethal Prostate Cancer: Is the Emergence of CRPC Inevitable?
To explain anti-neoplastic drug resistance, Goldie and Coldman applied principles first elaborated by Luria and Delbrück in classic studies of the resistance of bacteria to phage lysis to discriminate the contributions of spontaneous versus induced mutagenesis [62, 63]. Of course, all human cancer cells arise as a consequence of somatic genome errors, and most tend to show an increased propensity for genome instability. Cancer genome sequencing and other genome analyses have disclosed abundant base changes, insertions, deletions, amplifications, chromosome copy number changes, and DNA methylation differences in prostate cancers and other human cancers [64]. Early estimates hint that prostate cancer genomes contain on the order of 3,866 base changes, 108 rearrangements, and 5,408 differentially hypermethylated sequences [65, 66]. Ongoing genome instability can clearly result in cancer treatment resistance. Studies of cancer cell resistance to anti-metabolites and cytotoxic agents have consistently implicated a spontaneous mutation process of some sort as responsible for the emergence of resistant cancer cell clones among cells that were otherwise sensitive to the drugs. Such spontaneous mutation rates have been reported to be high as 1 in 104 per cell/generation in model studies [67]. For prostate cancer, both spontaneous and induced genome alterations probably contribute to disease progression to castration-resistance. The propensity for AR to recruit TOP2B to the regulatory regions of its target genes in prostate cancer cells and trigger directed chromosomal translocations may be a source of induced genome defects that can drive CRPC [14]. As an example, AR–TOP2B-associated DNA double strand breaks might conceivably promote AR amplification and AR over-expression. Prostate cancers have been proposed to evolve via rare widespread chromosomal rearrangement events termed “chromoplexy,” especially in cases showing AR-regulated fusion genes [68]. In contrast, spontaneous mutagenesis is likely responsible for the appearance of F876L-AR in men with CRPC treated with “second-generation” anti-androgens.
Genome-scale analyses of CRPC recovered at autopsy have consistently implicated a single lethal clone and its progeny as responsible for metastatic dissemination and ultimate life-threatening progression. However, ongoing genome instability in progeny of the lethal clone has been a consistent finding, suggesting that many, if not most, prostate cancers will be able to progress despite currently available treatment with androgen deprivation, inhibition of androgen synthesis, “second-generation” anti-androgens, and taxane chemotherapy, as well as to ultimately evade any future attempt at targeted therapy. In one study, genome copy number analysis showed greater similarity in losses and gains among metastatic cancer deposits in one case versus another, but distinct differences among the metastases in each case [69]. An analysis of somatic DNA hypermethylation changes capable of affecting gene expression delivered a similar result [70]. Intriguingly, in this study loss of cytosine methylation, evident in all metastatic prostate cancers, showed marked differences in every metastatic lesion, even within a single CRPC case, regardless of metastatic site. Hypomethylation appears to vary cell-to-cell in metastatic prostate cancer [67]. In addition to reducing the fidelity of suppression of normally silenced embryonic genes, like NY-ESO and others, this epigenetic instability may augment genetic instability in prostate cancer cells via activating retrotransposons and reducing chromatin barriers to repeat sequence recombination [70, 71].
Conclusions
The initial sensitivity of prostate cancers to androgen deprivation likely reflects a redirection of AR signaling from terminal differentiation toward maintenance of a malignant phenotype. This prostate cancer cell addiction to AR forms the basis for androgen deprivation therapy, the most widely used systemic treatment for advanced prostate cancer. Unfortunately, prostate cancers show enough genetic and epigenetic instability that disease progression to castration-resistance is inevitable. CPRC that has remained addicted to AR, particularly if the AR contains an intact LBD, is often amenable to treatment with androgen synthesis inhibitors and “second-generation” anti-androgens that selectively target the receptor LBD. CRPC that is driven by truncated AR encoded by splice variant AR transcripts, or that has escaped AR addiction, tends to be refractory to such drugs. AR-independent prostate cancers appear to be highly aggressive and difficult to treat with the current armamentarium of available anti-neoplastic drugs.
References
Gelmann EP. Molecular biology of the androgen receptor. J Clin Oncol. 2002;20(13):3001–15.
Lawrence MG, Lai J, Clements JA. Kallikreins on steroids: structure, function, and hormonal regulation of prostate-specific antigen and the extended kallikrein locus. Endocr Rev. 2010;31(4):407–46.
Lin B, Ferguson C, White JT, Wang S, Vessella R, True LD, Hood L, Nelson PS. Prostate-localized and androgen-dependent regulated expression of the membrane-bound serine protease TMPRSS2. Cancer Res. 1999;59(17):4180–4.
Nelson WG, Carter HB, DeWeese TL, Antonarakis ES, Eisenberger MA. Prostate cancer. In: Abeloff’s Clinical Oncology, Fifth Edition. Ed(s) Niederhuber JE, Armitage JO, Doroshow JH, Kastan MB, Tepper JE. Elsevier: Philadelphia, PA, 1653-1700, 2014. ISBN: 978-1-4557-2865-7.
Knudsen KE, Scher HI. Starving the addiction: new opportunities for durable suppression of AR signaling in prostate cancer. Clin Cancer Res. 2009;15(15):4792–8.
Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, Lee C, Montie JE, Shah RB, Pienta KJ, Rubin MA, Chinnaiyan AM. Recurrent fusions of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310(5748):644–8.
Mosquera JM, Mehra R, Regan MM, Perner S, Genega EM, Bueti G, Shah RB, Gaston S, Tomlins SA, Wei JT, Kearney MC, Johnson LA, Tang JM, Chinnaiyan AM, Rubin MA, Sanda MG. Prevalence of TMPRSS2-ERG fusion prostate cancer among men undergoing prostate biopsy in the United States. Clin Cancer Res. 2009;15(14):4706–11.
Rubin MA, Maher CA, Chinnaiyan AM. Common gene rearrangements in prostate cancer. J Clin Oncol. 2011;29(27):3659–68.
Klezovitch O, Risk M, Coleman I, Lucas JM, Null M, True LD, Nelson PS, Vasioukhin V. A causal role for ERG in neoplastic transformation of prostate epithelium. Proc Natl Acad Sci U S A. 2008;105(6):2105–10.
Tomlins SA, Laxman B, Varambally S, Cao X, Yu J, Helgeson BE, Cao Q, Prensner JR, Rubin MA, Shah RB, Mehra R, Chinnaiyan AM. Role of the TMPRSS2-ERG fusion gene in prostate cancer. Neoplasia. 2008;10(2):177–88.
Yu J, Yu J, Mani RS, Cao Q, Brenner CJ, Cao X, Wang X, Wu L, Li J, Hu M, Gong Y, Cheng H, Laxman B, Vellaichamy A, Shankar S, Li Y, Dhanasekaran SM, Morey R, Barrette T, Lonigro RJ, Tomlins SA, Varambally S, Qin ZS, Chinnaiyan AM. An integrated network of androgen receptor, polycomb, and TMPRSS2–ERG gene fusions in prostate cancer progression. Cancer Cell. 2010;17(5):443–54.
Carver BS, Chapinski C, Wongvipat J, Hieronymus H, Chen Y, Chandarlapaty S, Arora VK, Le C, Koutcher J, Scher H, Scardino PT, Rosen N, Sawyers CL. Reciprocal feedback regulation of PI3K and androgen receptor signaling in PTEN-deficient prostate cancer. Cancer Cell. 2011;19(5):575–86.
Chen Y, Chi P, Rockowitz S, Iaquinta PJ, Shamu T, Shukla S, Gao D, Sirota I, Carver BS, Wongvipat J, Scher HI, Zheng D, Sawyers CL. ETS factors reprogram the androgen receptor cistrome and prime prostate tumorigenesis in response to PTEN loss. Nat Med. 2013;19(8):1023–9.
Haffner MC, Aryee MJ, Toubaji A, Esopi DM, Albadine R, Gurel B, Isaacs WB, Bova GS, Liu W, Xu J, Meeker AK, Netto G, De Marzo AM, Nelson WG, Yegnasubramanian S. Androgen-induced TOP2B-mediated double-strand breaks and prostate cancer gene rearrangements. Nat Genet. 2010;42(8):668–75.
Weier C, Haffner MC, Mosbruger T, Esopi DM, Hicks J, Zheng Q, Fedor H, Isaacs WB, De Marzo AM, Nelson WG, Yegnasubramanian S. Nucleotide resolution of TMPRSS2 and ERG gene rearrangements in prostate cancer. J Pathol. 2013;230(2):174–83.
Li ZY, Liu DP, Liang CC. New insight into the molecular mechanisms of MLL-associated leukemia. Leukemia. 2005;19(2):183–90.
Lu NZ, Wardell SE, Burnstein KL, Defranco D, Fuller PJ, Giguere V, Hochberg RB, McKay L, Renoir JM, Weigel NL, Wilson EM, McDonnell DP, Cidlowski JA. International Union of Pharmacology. LXV. The pharmacology and classification of the nuclear receptor superfamily: glucocorticoid, mineralocorticoid, progesterone, and androgen receptors. Pharmacol Rev. 2006;58(4):782–97.
Beilin J, Ball EM, Favaloro JM, Zajac JD. Effect of the androgen receptor CAG repeat polymorphism on transcriptional activity: specificity in prostate and non-prostate cell lines. J Mol Endocrinol. 2000;25(1):85–96.
Giovannucci E, Stampfer MJ, Krithivas K, Brown M, Dahl D, Brufsky A, Talcott J, Hennekens CH, Kantoff PW. The CAG repeat within the androgen receptor gene and its relationship to prostate cancer. Proc Natl Acad Sci U S A. 1997;94(7):3320–3.
Shafi AA, Yen AE, Weigel NL. Androgen receptors in hormone-dependent and castration-resistant prostate cancer. Pharmacol Ther. 2013;140:223–38.
van de Wijngaart DJ, Dubbink HJ, van Royen ME, Trapman J, Jenster G. Androgen receptor co-regulators: recruitment via the co-activator groove. Mol Cell Endocrinol. 2012;352(1–2):57–69.
Bourguet W, Germain P, Gronemeyer H. Nuclear receptor ligand-binding domains: three-dimensional structures, molecular interactions and pharmacological implications. Trends Pharmacol Sci. 2000;21(10):381–8.
Wang Q, Li W, Zhang Y, Yuan X, Xu K, Yu J, Chen Z, Beroukhim R, Wang H, Lupien M, Wu T, Regan MM, Meyer CA, Carroll JS, Manrai AK, Jänne OA, Balk SP, Mehra R, Han B, Chinnaiyan AM, Rubin MA, True L, Fiorentino M, Fiore C, Loda M, Kantoff PW, Liu XS, Brown M. Androgen receptor regulates a distinct transcription program in androgen-independent prostate cancer. Cell. 2009;138(2):245–56.
Litvinov IV, Vander Griend DJ, Antony L, Dalrymple S, De Marzo AM, Drake CG, Isaacs JT. Androgen receptor as a licensing factor for DNA replication in androgen-sensitive prostate cancer cells. Proc Natl Acad Sci U S A. 2006;103(41):15085–90.
Kousteni S, Bellido T, Plotkin LI, O'Brien CA, Bodenner DL, Han L, Han K, DiGregorio GB, Katzenellenbogen JA, Katzenellenbogen BS, Roberson PK, Weinstein RS, Jilka RL, Manolagas SC. Nongenotropic, sex-nonspecific signaling through the estrogen or androgen receptors: dissociation from transcriptional activity. Cell. 2001;104(5):719–30.
Scher HI, Sawyers CL. Biology of progressive, castration-resistant prostate cancer: directed therapies targeting the androgen-receptor signaling axis. J Clin Oncol. 2005;23(32):8253–61.
Mostaghel EA, Page ST, Lin DW, et al. Intraprostatic androgens and androgen-regulated gene expression persist after testosterone suppression: therapeutic implications for castration-resistant prostate cancer. Cancer Res. 2007;67(10):5033–41.
Green SM, Mostaghel EA, Nelson PS. Androgen action and metabolism in prostate cancer. Mol Cell Endocrinol. 2012;360(1–2):3–13.
de Bono JS, Logothetis CJ, Molina A, et al. Abiraterone and increased survival in metastatic prostate cancer. N Engl J Med. 2011;364(21):1995–2005.
Vasaitis TS, Bruno RD, Njar VC. CYP17 inhibitors for prostate cancer therapy. J Steroid Biochem Mol Biol. 2011;125(1–2):23–31.
Kelly WK, Scher HI. Prostate specific antigen decline after antiandrogen withdrawal: the flutamide withdrawal syndrome. J Urol. 1993;149(3):607–9.
Taplin ME, Rajeshkumar B, Halabi S, Werner CP, Woda BA, Picus J, et al. Androgen receptor mutations in androgen-independent prostate cancer: Cancer and Leukemia Group B Study 9663. J Clin Oncol. 2003;21(14):2673–8.
Chen CD, Welsbie DS, Tran C, Baek SH, Chen R, Vessella R, et al. Molecular determinants of resistance to antiandrogen therapy. Nat Med. 2004;10(1):33–9.
Visakorpi T, Hyytinen E, Koivisto P, Tanner M, Keinänen R, Palmberg C, Palotie A, Tammela T, Isola J, Kallioniemi OP. In vivo amplification of the androgen receptor gene and progression of human prostate cancer. Nat Genet. 1995;9(4):401–6.
Waltering KK, Urbanucci A, Visakorpi T. Androgen receptor (AR) aberrations in castration-resistant prostate cancer. Mol Cell Endocrinol. 2012;360(1–2):38–43.
Tran C, Ouk S, Clegg NJ, Chen Y, Watson PA, Arora V, Wongvipat J, Smith-Jones PM, Yoo D, Kwon A, Wasielewska T, Welsbie D, Chen CD, Higano CS, Beer TM, Hung DT, Scher HI, Jung ME, Sawyers CL. Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science. 2009;324(5928):787–90.
Clegg NJ, Wongvipat J, Joseph JD, Tran C, Ouk S, Dilhas A, Chen Y, Grillot K, Bischoff ED, Cai L, Aparicio A, Dorow S, Arora V, Shao G, Qian J, Zhao H, Yang G, Cao C, Sensintaffar J, Wasielewska T, Herbert MR, Bonnefous C, Darimont B, Scher HI, Smith-Jones P, Klang M, Smith ND, De Stanchina E, Wu N, Ouerfelli O, Rix PJ, Heyman RA, Jung ME, Sawyers CL, Hager JH. ARN-509: a novel antiandrogen for prostate cancer treatment. Cancer Res. 2012;72(6):1494–503.
Scher HI, Fizazi K, Saad F, Taplin ME, Sternberg CN, Miller K, de Wit R, Mulders P, Chi KN, Shore ND, Armstrong AJ, Flaig TW, Fléchon A, Mainwaring P, Fleming M, Hainsworth JD, Hirmand M, Selby B, Seely L, de Bono JS, AFFIRM Investigators. Increased survival with enzalutamide in prostate cancer after chemotherapy. N Engl J Med. 2012;367(13):1187–97.
Joseph JD, Lu N, Qian J, Sensintaffar J, Shao G, Brigham D, et al. A clinically relevant androgen receptor mutation confers resistance to 2nd generation anti-androgens enzalutamide and ARN-509. Cancer Discov. 2013;3:1020–9.
Korpala M, Korna JM, Gaob X, Rakiecc DP, Ruddyc DA, Doshia S, et al. A novel mutation in androgen receptor confers genetic and phenotypic resistance to MDV3100 (enzalutamide). Cancer Discov. 2013;3:1030–43.
Balbas MD, Evans MJ, Hosfield DJ, Wongvipat J, Arora VK, Watson PA, et al. Overcoming mutation-based resistance to antiandrogens with rational drug design. eLife. 2013;2:e00499.
Watson PA, Chen YF, Balbas MD, Wongvipat J, Socci ND, Viale A, Kim K, Sawyers CL. Constitutively active androgen receptor splice variants expressed in castration-resistant prostate cancer require full-length androgen receptor. Proc Natl Acad Sci U S A. 2010;107(39):16759–65.
Sun S, Sprenger CC, Vessella RL, Haugk K, Soriano K, Mostaghel EA, Page ST, Coleman IM, Nguyen HM, Sun H, Nelson PS, Plymate SR. Castration resistance in human prostate cancer is conferred by a frequently occurring androgen receptor splice variant. J Clin Invest. 2010;120(8):2715–30.
Hu R, Dunn TA, Wei S, Isharwal S, Veltri RW, Humphreys E, Han M, Partin AW, Vessella RL, Isaacs WB, Bova GS, Luo J. Ligand-independent androgen receptor variants derived from splicing of cryptic exons signify hormone-refractory prostate cancer. Cancer Res. 2009;69(1):16–22.
Guo Z, Yang X, Sun F, Jiang R, Linn DE, Chen H, Chen H, Kong X, Melamed J, Tepper CG, Kung HJ, Brodie AM, Edwards J, Qiu Y. A novel androgen receptor splice variant is up-regulated during prostate cancer progression and promotes androgen depletion-resistant growth. Cancer Res. 2009;69(6):2305–13.
Dehm SM, Schmidt LJ, Heemers HV, Vessella RL, Tindall DJ. Splicing of a novel androgen receptor exon generates a constitutively active androgen receptor that mediates prostate cancer therapy resistance. Cancer Res. 2008;68(13):5469–77.
Nyquist MD, Dehm SM. Interplay between genomic alterations and androgen receptor signaling during prostate cancer development and progression. Horm Cancer. 2013;4(2):61–9.
Liu LL, Xie N, Sun S, Plymate S, Mostaghel E, Dong X. Mechanisms of the androgen receptor splicing in prostate cancer cells. Oncogene. 2014;33(24):3140–50. doi:10.1038/onc.2013.284. Epub 2013 Jul 15.
Hörnberg E, Ylitalo EB, Crnalic S, Antti H, Stattin P, Widmark A, Bergh A, Wikström P. Expression of androgen receptor splice variants in prostate cancer bone metastases is associated with castration-resistance and short survival. PLoS One. 2011;6(4):e19059.
Hu R, Lu C, Mostaghel EA, Yegnasubramanian S, Gurel M, Tannahill C, Edwards J, Isaacs WB, Nelson PS, Bluemn E, Plymate SR, Luo J. Distinct transcriptional programs mediated by the ligand-dependent full-length androgen receptor and its splice variants in castration-resistant prostate cancer. Cancer Res. 2012;72(14):3457–62.
Li Y, Chan SC, Brand LJ, Hwang TH, Silverstein KA, Dehm SM. Androgen receptor splice variants mediate enzalutamide resistance in castration-resistant prostate cancer cell lines. Cancer Res. 2013;73(2):483–9.
Nelson PS. Molecular states underlying androgen receptor activation: a framework for therapeutics targeting androgen signaling in prostate cancer. J Clin Oncol. 2012;30:644–6.
Hirano D, Okada Y, Minei S, Takimoto Y, Nemoto N. Neuroendocrine differentiation in hormone refractory prostate cancer following androgen deprivation therapy. Eur Urol. 2004;45(5):586–92.
Schelling LA, Williamson SR, Zhang S, Yao JL, Wang M, Huang J, Montironi R, Lopez-Beltran A, Emerson RE, Idrees MT, Osunkoya AO, Man YG, Maclennan GT, Baldridge LA, Compérat E, Cheng L. Frequent TMPRSS2-ERG rearrangement in prostatic small cell carcinoma detected by fluorescence in situ hybridization: the superiority of fluorescence in situ hybridization over ERG immunohistochemistry. Hum Pathol. 2013;44:2227–33.
Beltran H, Rickman DS, Park K, Chae SS, Sboner A, MacDonald TY, Wang Y, Sheikh KL, Terry S, Tagawa ST, Dhir R, Nelson JB, de la Taille A, Allory Y, Gerstein MB, Perner S, Pienta KJ, Chinnaiyan AM, Wang Y, Collins CC, Gleave ME, Demichelis F, Nanus DM, Rubin MA. Molecular characterization of neuroendocrine prostate cancer and identification of new drug targets. Cancer Discov. 2011;1(6):487–95.
Aparicio AM, Harzstark AL, Corn PG, Wen S, Araujo JC, Tu SM, et al. Platinum-based chemotherapy for variant castrate-resistant prostate cancer. Clin Cancer Res. 2013;19(13):3621–30.
Beltran H, Tagawa ST, Park K, MacDonald T, Milowsky MI, Mosquera JM, Rubin MA, Nanus DM. Challenges in recognizing treatment-related neuroendocrine prostate cancer. J Clin Oncol. 2012;30(36):e386–9.
Logothetis CJ, Gallick GE, Maity SN, Kim J, Aparicio A, Efstathiou E, Lin SH. Molecular classification of prostate cancer progression: foundation for marker-driven treatment of prostate cancer. Cancer Discov. 2013;3(8):849–61.
Dorkin TJ, et al. FGF8 over-expression in prostate cancer is associated with decreased patient survival and persists in androgen independent disease. Oncogene. 1999;18:2755–61.
Wu YM, Su F, Kalyana-Sundaram S, Khazanov N, Ateeq B, Cao X, Lonigro RJ, Vats P, Wang R, Lin SF, Cheng AJ, Kunju LP, Siddiqui J, Tomlins SA, Wyngaard P, Sadis S, Roychowdhury S, Hussain MH, Feng FY, Zalupski MM, Talpaz M, Pienta KJ, Rhodes DR, Robinson DR, Chinnaiyan AM. Identification of targetable FGFR gene fusions in diverse cancers. Cancer Discov. 2013;3(6):636–47.
Schoenborn JR, Nelson P, Fang M. Genomic profiling defines subtypes of prostate cancer with the potential for therapeutic stratification. Clin Cancer Res. 2013;19:4058–66.
Luria SE, Delbruck M. Mutations of bacteria from virus sensitivity to virus resistance. Genetics. 1943;28(6):491–511.
Goldie JH, Coldman AJ. A mathematic model for relating the drug sensitivity of tumors to their spontaneous mutation rate. Cancer Treat Rep. 1979;63(11–12):1727–33.
Vogelstein B, Papadopoulos N, Velculescu VE, Zhou S, Diaz Jr LA, Kinzler KW. Cancer genome landscapes. Science. 2013;339(6127):1546–58.
Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary prostate cancer. Nature. 2011;470(7333):214–20.
Berger MF, Lawrence MS, Demichelis F, Drier Y, Cibulskis K, Sivachenko AY, Sboner A, Esgueva R, Pflueger D, Sougnez C, Onofrio R, Carter SL, Park K, Habegger L, Ambrogio L, Fennell T, Parkin M, Saksena G, Voet D, Ramos AH, Pugh TJ, Wilkinson J, Fisher S, Winckler W, Mahan S, Ardlie K, Baldwin J, Simons JW, Kitabayashi N, MacDonald TY, Kantoff PW, Chin L, Gabriel SB, Gerstein MB, Golub TR, Meyerson M, Tewari A, Lander ES, Getz G, Rubin MA, Garraway LA. The genomic complexity of primary prostate cancer. Nature. 2011;470(7333):214–20.
Yegnasubramanian S, Wu Z, Haffner MC, Esopi D, Aryee MJ, Badrinath R, He TL, Morgan JD, Carvalho B, Zheng Q, De Marzo AM, Irizarry RA, Nelson WG. Chromosome-wide mapping of DNA methylation patterns in normal and malignant prostate cells reveals pervasive methylation of gene-associated and conserved intergenic sequences. BMC Genomics. 2011;12:313.
Tlsty TD, Margolin BH, Lum K. Differences in the rates of gene amplification in nontumorigenic and tumorigenic cell lines as measured by Luria–Delbruck fluctuation analysis. Proc Natl Acad Sci U S A. 1989;86(23):9441–5.
Baca SC, Prandi D, Lawrence MS, Mosquera JM, Romanel A, Drier Y, Park K, Kitabayashi N, MacDonald TY, Ghandi M, Van Allen E, Kryukov GV, Sboner A, Theurillat JP, Soong TD, Nickerson E, Auclair D, Tewari A, Beltran H, Onofrio RC, Boysen G, Guiducci C, Barbieri CE, Cibulskis K, Sivachenko A, Carter SL, Saksena G, Voet D, Ramos AH, Winckler W, Cipicchio M, Ardlie K, Kantoff PW, Berger MF, Gabriel SB, Golub TR, Meyerson M, Lander ES, Elemento O, Getz G, Demichelis F, Rubin MA, Garraway LA. Punctuated evolution of prostate cancer genomes. Cell. 2013;153(3):666–77.
Liu W, Laitinen S, Khan S, Vihinen M, Kowalski J, Yu G, Chen L, Ewing CM, Eisenberger MA, Carducci MA, Nelson WG, Yegnasubramanian S, Luo J, Wang Y, Xu J, Isaacs WB, Visakorpi T, Bova GS. Copy number analysis indicates monoclonal origin of lethal metastatic prostate cancer. Nat Med. 2009;15(5):559–65.
Aryee MJ, Liu W, Engelmann JC, Nuhn P, Gurel M, Haffner MC, Esopi D, Irizarry RA, Getzenberg RH, Nelson WG, Luo J, Xu J, Isaacs WB, Bova GS, Yegnasubramanian S. DNA methylation alterations exhibit intraindividual stability and interindividual heterogeneity in prostate cancer metastases. Sci Transl Med. 2013;5(169):169ra10.
Yegnasubramanian S, Haffner MC, Zhang Y, Gurel B, Cornish TC, Wu Z, Irizarry RA, Morgan J, Hicks J, DeWeese TL, Isaacs WB, Bova GS, De Marzo AM, Nelson WG. DNA hypomethylation arises later in prostate cancer progression than CpG island hypermethylation and contributes to metastatic tumor heterogeneity. Cancer Res. 2008;68(21):8954–67.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Nelson, W.G., Pienta, K.J. (2014). Molecular Mechanisms of Prostate Cancer Progression After Castration. In: Saad, F., Eisenberger, M. (eds) Management of Castration Resistant Prostate Cancer. Current Clinical Urology. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-1176-9_3
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1176-9_3
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-1175-2
Online ISBN: 978-1-4939-1176-9
eBook Packages: MedicineMedicine (R0)