Abstract
Cells coordinate chaperones at the exit site of the ribosome. Albeit the types and mechanisms of ribosome-associated chaperones differ in the three kingdoms of life, they all share the ability to protect nascent polypeptides from off pathways such as aggregation and degradation and, at least in some cases, support initial folding steps of newly synthesized proteins. Recent progress was made in understanding the nascent interactome of these ribosome-associated chaperones. While the bacteria-specific chaperone trigger factor (TF) binds to almost every nascent polypeptide made by ribosomes except for membrane proteins, the substrate pool of the two eukaryotic ribosome-associated chaperone systems, nascent polypeptide-associated complex (NAC) and ribosome-associated complex (RAC), is more distinct.
Interestingly, there is culminating evidence that these chaperones also display important functions off the ribosome, e.g., in the biogenesis of ribosomal subunits and in protein aggregation under proteotoxic stress conditions. In this chapter, we will discuss the functions of these chaperones with regard to their broad substrate pools.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Trigger factor
- Nascent polypeptide-associated complex (NAC)
- Hsp70
- Hsp40
- RAC
- De novo protein folding
- Ribosome biogenesis
- Proteotoxic stress
1 Introduction: Ribosome-Associated Chaperones in De novo Folding
Directly upon their synthesis by the ribosome, proteins have to fold into their unique three-dimensional structure in order to become biologically active. The folding of proteins is problematic since hydrophobic residues of the unfolded polypeptide chain are accessible, which enhances the probability that the newly synthesized protein follows an unproductive off pathway leading to misfolding and aggregation [1]. As protein misfolding and aggregation represent the hallmarks of several neurodegenerative diseases, it is of particular importance to understand the mechanisms by which proteins acquire and maintain their structure under cellular conditions. To accomplish folding and prevent off pathways, newly synthesized polypeptides interact with at least one cytosolic chaperone that prevents inappropriate inter- and intramolecular interactions, and thus promotes the folding into the native state. Among these chaperones, the ribosome-associated ones interact with nascent polypeptides while they are still attached to the peptidyl-transferase center of the ribosome (Figs. 2.1 and 2.2a) [2–4].
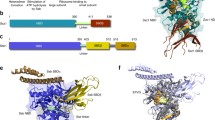
a E. coli trigger factor. The N-domain (red) contains the ribosome-binding motif (40-GFRxGxxP-49). It is located in a loop region between two α-helices, the N-domain is connected to the PPIase domain (green) via an extended linker. The C-domain (blue) is located in the center of the folded molecule and forms two arm-like protrusions. The N-domain and both arms of the C-domain together form a cavity for nascent polypeptide chains. b The nascent polypeptide-associated complex (NAC). The α-subunit of NAC (encoded in yeast by the EGD2 gene) consists of a NAC-domain in the N-terminal region and an ubiquitin-associated (UBA) domain at its C-terminus. In yeast two alternative β-subunits (β and β′) exist, which are either encoded by the genes EGD1 or BTT1, respectively. The β-subunits of NAC contain a conserved ribosome-binding motif in the N-terminus and a central NAC domain. Dimerization of the NAC subunits involves the NAC domains of both subunits. α-NAC (blue) and β-NAC (orange) form a stable heterodimer. The UBA domain is derived from the crystal structure of archaeal NAC (PDB 1TR8) and was modeled on the human NAC domain heterodimer PDB 3LKX, aa 84–136 of α-NAC (NACA) and aa 97–162 of β-NAC (BTF3 isoform). Broken lines indicate unresolved parts of the molecule. c The ribosome-asscoiated heat shock protein (Hsp)70/40 system in eukaryotes. The yeast ribosome-associated chaperone system consists of the Hsp70s Ssb, Ssz, and the Hsp40 Zuo. Ssb1 and Ssz contain an N-terminal nucleotide-binding domain (NBD, violet) and a C-terminal substrate-binding domain (SBD, dark green). The SBD of Ssz is shorter compared to canonical Hsp70s. Like all Hsp40 co-chaperones Zuo contains a J-domain (light green) required for stimulation of the adenosine triphosphatase (ATPase) activity of its Hsp70 partner. In addition, Zuo contains a charged region (blue) in its C-terminus, which is involved in ribosome binding. The N-terminus (N, orange) of Zuo is predicted to be unstructured. Ssz and Zuo form the stable heterodimeric ribosome-associated complex (RAC). A schematic model of RAC based on small-angle X-ray scattering (SAXS) analysis of yeast RAC (on the right side, shown in light grey) reveals that the complex forms an elongated structure that was divided into body, neck and head. Most of the body is composed of Ssz, whereas Zuo forms the neck and the head. The head region of Zuo was shown to contact the ribosome near the tunnel exit. Lower panel: In contrast, in human, no Ssb homolog is found while RAC is conserved. The RAC complex is formed by the Hsp40 MPP11 and Hsp70L1. MPP11 contains two SANT domains (pink) at its C-terminus. The functions of the SANT domains are still unknown
a Ribosome-associated chaperones bind to ribosomes (grey) in close proximity to the ribosomal tunnel exit and interact with nascent polypeptide chains (yellow). In bacteria, only trigger factor (orange) binds to the ribosome and interacts with the growing nascent polypeptide. In eukaryotes, two chaperone systems are found in association with the ribosome. On the one hand, the heterodimeric nascent polypeptide-associated complex (NAC) complex that consists of a α- (dark blue) and β- (light blue) subunit binds to the ribosome via its β-subunit. Additionally, a system consisting of heat shock protein (Hsp)70 and Hsp40 family members binds to ribosomes and interacts with nascent chains. In yeast this system comprises the Hsp70 Ssb (pink) as well as the ribosome-associated complex (RAC) complex, which is formed by the Hsp40 Zuo (light green) and the Hsp70 Ssz (violet). Mammalian RAC (mRAC) is formed by the Hsp40 MPP11 and Hsp70L1. As Ssb is restricted to fungi, cytosolic Hsp70s (red) are recruited to the nascent chain by mRAC. b The table summarizes the co-translational substrate specificities and the substrate pool of trigger factor from E. coli as well as Ssb and NAC from S. cerevisiae. The substrate specificities of trigger factor were taken from: [36]; the substrate pool from: [27]. The co-translational interactome of Ssb was analyzed in [92] and that of NAC in [79]
Binding of chaperones to nascent polypeptides can have several effects. The unfolded polypeptide is protected from being degraded or from incorrect contact with other molecules that may lead to aggregation. Moreover, for polypeptides that start their folding program co-translationally, chaperone binding might support very early folding. Beyond the interaction of newly synthesized proteins with ribosome-attached chaperones, additionally other cytosolic chaperones can act during later stages of translation on elongated nascent chains or after release of the polypeptides from the ribosomes. The latter mainly belong to the heat shock protein (Hsp)70/40 and the Hsp60/10 chaperone families. Examples are the DnaK/DnaJ (Hsp70/Hsp40) and GroEL/GroES (Hsp60/Hsp10) systems in the cytosol of E. coli cells [3, 5]. Together with ribosome-asssociated chaperones they form a robust network that promotes the de novo folding of newly synthesized polypeptides and prevents off pathways such as aggregation or degradation early in the life of a new protein [4].
Ribosome-associated chaperones are found in every cell but differ significantly among the different kingdoms of life with regard to their number and type. Whereas prokaryotes have only one ribosome-associated chaperone which is called trigger factor (TF) [6–9], eukaryotic ribosomes coordinate two TF-unrelated chaperone systems at the ribosome (Fig. 2.2a). These systems are the highly conserved nascent polypeptide-associated complex (NAC) and an arrangement of specialized Hsp70 and Hsp40 chaperones that includes a heterodimer called ribosome-associated complex (RAC; Fig. 2.2a) [4, 10]. In yeast, the RAC system has a third ribosome-associated partner, a Hsp70 chaperone called Ssb [11] (Fig. 2.2a) .
In the following section, the functions and structures of these ribosome-associated chaperones will be discussed together with their respective substrates.
2 The Prokaryotic Ribosome-Associated Chaperone Trigger Factor
The only known chaperone of bacterial cells with a direct binding site on ribosomes is TF . In E.coli, TF consists of 432 amino acids and has a molecular weight of 48 kDa. It is a three-domain protein with an N-terminal ribosome-binding domain, a middle domain that displays peptidyl-prolyl cis/trans isomerase (PPIase) activity and a C-terminal domain [7, 12] (Fig. 2.1a). Its crystal structure revealed that TF adopts an extended three-dimensional conformation with an unusual domain arrangement [9]. Although distant in the amino acid sequence, the N- and C-terminal domains are located adjacent to each other in the three-dimensional structure to form a cradle-like structure (Fig. 2.1a). The C-terminal domain of TF is located in the center of the molecule and forms two arm-like protrusions [9] . The N-terminal domain contains a signature motif (42-GFRxGxxP-50) that is located in an exposed loop region [9] and binds to the ribosomal protein L23 (Fig. 2.1a). Mutation of either the signature motif or a conserved surface-exposed residue within the ribosomal protein L23 strongly impairs ribosome binding of TF and its activity on nascent polypeptides [13]. The N-terminus is connected via a long linker to the second domain, the PPIase domain, which is located at the opposite end of the molecule [9] (Fig. 2.1a). This domain catalyzes prolyl cis/trans isomerization in peptides in vitro and presumably slows down folding processes or acts as auxiliary site to assist the folding of substrate proteins [14–16]. However, the in vivo relevance of this domain and its enzymatic activity is still not well-understood [17] .
Crystallization of the N-terminal fragment of E. coli TF together with the 50S large ribosomal subunit from Haloarcula marismortui allowed the superposition of full-length TF with ribosomes and paved the way for understanding how this chaperone functions on ribosomes [9]. TF binds to the ribosomal protein L23 and hunches with its extended cradle-like structure over the ribosomal tunnel exit site (Fig. 2.2a). It was suggested that the cradle-like structure provides a shielded environment for the emerging nascent polypeptides and thus supports co-translational folding events [9]. The interior of the cavity formed by the TF N- and C-terminal domains contains hydrophobic as well as hydrophilic areas. Indeed, multiple potential substrate-binding sites within this cradle have been suggested [18, 19]. This structure might enable TF to bind a large variety of substrates .
The ribosome-binding activity of TF has been extensively characterized. It cycles on and off the ribosomes and binds as a monomer to the translation machinery [14, 20, 21]. TF is present in a two- to threefold molar excess over ribosomes and thus is bound to most cytosolic ribosomes. Unbound TF diffuses freely in the cytosol, probably as a homodimer. The in vivo relevance of this dimerization is unclear but dimer formation may represent a storage form of this chaperone to prevent its cradle from unspecific associations [22, 23]. Although TF binds to non-translating ribosomes with a KD of approximately 1 µM and a mean residence time of 10–15 s, the presence of a nascent chain increases the affinity of TF for ribosomes up to 30-fold [20, 24–26]. It could be shown by cross-linking studies combined with cryo-electron microscopy structures that TF interacts with very short nascent chains as soon as they emerge from the ribosomal exit site in vitro [18]. In vivo, however, recruitment of TF to ribosome-nascent chain complexes is delayed until the polypeptides reach a length of 100 amino acids [27]. TF binding to nascent substrates is controlled by processing enzymes that act first on the N-terminus of the newly synthesized protein. It was demonstrated recently that peptide deformylase associates with nascent chains as soon as they emerge from the ribosomal exit tunnel, followed by methionine aminopeptidase [28]. Both enzymes prevent the premature recruitment of TF [27, 29]. Most likely TF can also stay bound to a subset of nascent polypeptides after their release from the ribosome [30] .
Initially, TF was discovered as a cytosolic protein involved in the translocation of the outer membrane protein pro-OmpA across the plasma membrane as it has the ability to promote the insertion of chemically denatured pro-OmpA into membrane vesicles [31]. However, TF is not essential for viability of E. coli and deletion of its gene tig does not cause any growth defect albeit the heat shock response is induced leading to enhanced levels of chaperones and proteases [32, 33]. First evidence that TF fulfills chaperone function in vivo came from the finding that the simultaneous deletion of TF and the cytosolic Hsp70 chaperone DnaK is synthetically lethal at temperatures above 30 °C [32, 34]. Cells with low levels of DnaK and without TF are viable but show slow growth and massive aggregation of several hundreds of cytosolic proteins, especially large-sized proteins and components of protein complexes . Interestingly, TF and DnaK show some overlap in their substrate specificity. Both chaperones interact with peptide segments of an unfolded protein that has a high mean hydrophobicity and an overall positive net charge [35, 36] (Fig. 2.2b). Taken together, these findings suggest that the cytosolic Hsp70 DnaK and the ribosome-associated chaperone TF act on a similar substrate spectrum and thus are able to cooperate in the de novo folding of newly synthesized proteins .
The mechanism by which the non-adenosine triphosphate (non-ATP)-consuming TF promotes de novo folding of newly synthesized proteins is still not fully understood. On the one hand, the cavity formed by the N- and C-terminal domains could provide a folding chamber that protects the emerging nascent chain from unfavorable interactions while translation proceeds. In agreement with these findings it could be demonstrated that TF protects nascent proteins from proteolytic digestion in vitro [37, 38]. Additionally, it was shown recently that TF stimulated native folding in constructs of repeated maltose-binding protein (MBP) domains by protecting partially folded domains from distant interactions that produce stable misfolded states [39]. On the other hand, it was proposed that TF delays the folding of a nascent polypeptide until sufficient sequence information (encoded in the C-terminal area of the synthesized polypeptide chain) is available outside of the ribosome to allow productive folding [35, 38]. Indeed, recent data suggest that TF can unfold preexisting folded states to prevent and revert premature folding, thus limiting the formation of misfolded intermediate states during protein synthesis [8]. In the current model, first the ribosome itself limits the conformational freedom of the newly synthesized polypeptide chains. As translation proceeds and the chain lengthens the influence of the ribosome, in particular on the N-terminal regions, decreases and the risk to form misfolded intermediates increases. Therefore, TF binds to nascent polypeptides of a length of 100 amino acids in repeated binding and release cycles. It limits conformational sampling and folding more efficiently than the ribosome and thereby prevents folding intermediates [8] . TF is not only able to block folding but also unfolds preformed segments in order to give the nascent chain a new opportunity for productive folding. After release from the ribosome, some nascent chains that do not need further assistance acquire their native structure after dissociation of TF. In contrast, other proteins stay associated with TF and are transferred to downstream chaperone systems, in particular to the DnaK/DnaJ/GrpE system or to GroEL/ES.
Very recently, the nascent interactome of ribosome-bound TF was identified using an elegant approach: selective ribosome profiling [27, 40]. For this technique, actively translating ribosomes are isolated; the messenger itochondrial ribonucleic acid (mRNA) bound to them is converted into deoxyribonucleic acid (DNA) and subsequently analyzed by high-throughput sequencing. To gain insights into which nascent polypeptides are bound by TF, only those ribosomes with bound TF were isolated and the mRNA was analyzed. TF was fused with an affinity purification tag, ribosomes were isolated, TF binding to the nascent polypeptides was stabilized by cross-linking, and thus specifically the ribosome-nascent-chain complexes containing TF could be isolated by affinity purification [27] .
The data revealed that the medium average length at which TF engaged a polypeptide was 112 amino acids, half of all nascent chains being bound within ± 20 amino acids of this position. Nascent chains are generally not engaged by more than one TF molecule. Furthermore, the data revealed that TF interacts with all nascent chains in E. coli except for those that localize to the plasma membrane (Fig. 2.2b). Membrane proteins are poorly bound by TF. Interestingly, outer membrane β-barrel proteins (OMPs) represent the strongest TF interactors (Fig. 2.2b). For example, the five best-characterized OMPs (LamB, LptD, OmpA, OmpC, and OmpF) were found among the 25 strongest TF-interacting nascent polypeptides. This finding is in agreement with very early data, as TF was initially discovered by its ability to promote insertion of pro-OmpA into membranes [31]. To prove the in vivo relevance of this finding the authors analyzed the protein content of the outer membrane fraction of wild type E. coli cells and compared it with those lacking TF using stable isotope labeling with amino acids in cell culture (SILAC) combined with mass spectrometry. The data revealed that in cells lacking TF, 60 % of all detected proteins were found in lower amounts than in the wild type, whereas no proteins were found in higher amounts. In agreement with this result, it was shown that loss of TF causes enhanced sensitivity of E. coli to SDS/EDTA (sodium dodecyl sulfate/ethylenediamine tetraacidic acid)treatment or vancomycin, which suggests defects in the biogenesis of outer membrane proteins. In sum, these data clearly suggest that TF not only plays an important role in the folding of cytosolic proteins but is also crucial for the folding-competent conformation of outer membrane proteins during their biogenesis [27] .
3 The Nascent Polypeptide-Associated Complex (NAC)
NAC is a heterodimeric complex that associates with ribosomes likely in a 1:1 stoichiometry [41, 42] (Fig. 2.2a) . It is highly conserved among eukaryotes and consists of one α- and one β-subunit, referred to as α-NAC and β-NAC (Fig. 2.1b). NAC is not found in bacteria but in archaea, where it is built by an α-NAC homodimer [43, 44].
Both subunits of heterodimeric NAC were shown to contact nascent polypeptide chains [42, 45] while only β-NAC interacts with the ribosome and mediates ribosome binding of the complex [43, 46] (Fig. 2.2a). NAC is present in equimolar concentration relative to ribosomes and binds to them irrespective of their translation status [41]. In yeast, NAC is encoded by three genes. The α-NAC subunit is encoded by EGD2 while EGD1 as well as BTT1 both encode for β-subunits. The gene product of EGD1 is referred to as β-NAC and the one of BTT1 as β′-NAC (Fig. 2.1b). Both β-subunits are able to form a complex with the α-subunit. However, the β′-NAC version is approximately 100 times less expressed than the β-version [44, 47]. Thus, the most abundant NAC type is formed by α β in yeast cells.
Both subunits of NAC share a homologous structural element, the so-called NAC-domain. These domains are responsible for the dimerization of NAC into the heterodimeric complex (Fig. 2.1b). The NAC-domain is located in the N-terminal part of α-NAC and in the central domain of β-NAC. Crystal structures of parts of the human complex give insights into the molecular interactions governing NAC complex formation [48, 49]. They reveal a handshake interaction of the two NAC-domains consisting of a six-stranded β-barrel that is stabilized by hydrophobic contacts between conserved residues [49, 50]. Additionally, α-NAC contains an ubiquitin-associated (UBA) domain [50] (Fig. 2.1b). UBA domains were described to bind mono- and polyubiquitin but also mediate other protein-protein interactions [51]. They are commonly found in factors dedicated to ubiquitin-dependent protein degradation or in components of signal transduction pathways [51–54]. However, the function of the UBA domain of α-NAC remains elusive .
There is no structure available of NAC in complex with the ribosome and thus its orientation on the ribosome and relative to the exit site for nascent polypeptides is unknown. Different physical contact points for NAC are under debate. Based on cross-linking data various anchor points have been suggested: either Rpl25, the L23 homolog that coordinates TF in bacteria [55] or Rpl31 [56, 57] which is also the main interaction site for RAC (see below). Both Rpl25 and Rpl31 are located directly at the ribosomal exit site and would position NAC in a way that favors its interaction with nascent polypeptides. Previous work suggested that NAC binds to Rpl25 via a conserved motif (RRK-(X)n-KK, amino acids 23–31 located in a loop region between two α-helices in the N-terminus of β-NAC (Fig. 2.1b). Upon mutation of the amino acid residues arginine arginine lysine RRK (amino acids 23–31) to alanine alanine alanine (AAA) ribosome binding of NAC was abolished both in vivo and in vitro [55] .
Despite the evolutionary conservation of NAC in eukaryotes, its in vivo function is still rather obscure. Originally, NAC was discovered in 1994 by Wiedman and coworkers and described as a translocation and sorting factor [42]. Based on in vitro data, they suggested that NAC interacts with ribosomes and nascent polypeptides in order to prevent mislocalization of ribosomes translating cytosolic proteins to the endoplasmic reticulum (ER) [42–46, 57–64]. However, in vivo evidence for such a function is missing till today. Another study reported a correlation between altered NAC levels and neurodegenerative diseases [65]. β-NAC was also described as a target for caspases [66] and an apoptosis-suppressing activity of NAC was reported [67]. Moreover, functions as a transcriptional regulator were described for individual NAC subunits [68–71] .
The deletion of the NAC-encoding genes has no effect on the growth or viability of yeast cells. However, β-NAC knockout cells (egd1∆btt1∆) where only the α-NAC subunit is expressed show a slight growth defect at 37 °C [44]. In contrast, there is an embryonic lethality of NAC mutants in C. elegans [67], D. melanogaster [72], and mice [73] indicating that this complex fulfills essential functions in higher eukaryotes.
Since NAC binds to ribosomes and interacts with nascent polypeptides, a chaperone-like function of NAC was proposed [4, 74–76]. First evidence for such a role was obtained by a recent study showing that NAC cooperates with the Ssb-RAC chaperone system in yeast [77] . It could be demonstrated that the absence of NAC enhances the sensitivity of ssb∆ cells towards drugs that cause misreading during translation or folding stress. Cells lacking Ssb and NAC show aggregation of newly synthesized polypeptides and defects in translation [77]. Another study showed that NAC is a central component of the protein homeostasis network in the metazoan animal model C. elegans [78]. Under non-stress conditions NAC is associated with the ribosome where it supports protein folding as well as translation activity. However, NAC seems to fulfill a dual function that becomes evident under stress conditions: Upon proteotoxic stress, NAC interacts with protein aggregates and thereby becomes discharged from its ribosomal housekeeping function . The localization of NAC to protein aggregates induced by heat shock conditions is important for the fast resolubilization of these aggregates after the stress. Likewise, the sequestration of NAC from ribosomes to protein aggregates causes a strong decrease in the cytosolic pool of actively translating polysomes. It is suggested that a decrease of translation is beneficial under stress conditions that cause protein misfolding and aggregation since the influx of newly synthesized and unfolded proteins is reduced. This, in turn, decreases the overall folding load for chaperones and allows the chaperone network to act on the remodeling of existing aggregates. Thus, NAC has crucial housekeeping and stress reducing functions in metazoans. It acts as a modulator of protein synthesis to establish a regulatory feedback mechanism that adjusts translational activity to the cellular protein folding load [78] .
A study by Frydman and co-workers recently identified the nascent interactome of NAC in yeast [79]. They expressed individual tandem-affinity-purification (TAP)-tagged NAC subunits to purify the NAC-ribosome-nascent chain complexes (NAC-RNCs) together with the corresponding mRNA. The mRNAs were identified by DNA microarray hybridization. Using this strategy, the authors were able to show that the different subunits of yeast NAC interact with distinct sets of nascent polypeptides, but virtually every nascent protein on ribosomes contacts at least one NAC subunit. In other words, every protein that is translated in a yeast cell interacts co-translationally with NAC. According to these data, β′-NAC associates with ribosomes translating mRNAs that encode proteins with high intrinsic disorder and low hydrophobicity as well as mitochondrial or ribosomal proteins. The substrates of β-NAC and α-NAC showed a large overlap and were more distinct from those of β′-NAC. For example, hydrophobic proteins and metabolic enzymes were highly enriched among their interactors [79] (Fig. 2.2b) .
As mentioned above, a role for NAC in co-translational protein targeting has been discussed since decades [42–46, 57–64]. Therefore, the authors also analyzed the interplay between the targeting factor SRP (signal recognition particle) and NAC. Originally, it was proposed that NAC and SRP compete for ribosome binding and that a nascent polypeptide can only interact with one of them at the same time. In contrast, this study indicates that both factors can bind simultaneously to one ribosome and the presence of NAC does not prevent the interaction of nascent chains efficiently with SRP in yeast. In the absence of NAC, however, a subset of the nascent secretory proteins was unable to interact with SRP, whereas on the other hand, increased false contacts between SRP and cytosolic proteins were detected which are no SRP substrates [79] .
In addition, NAC plays a role in protecting nascent chains from premature and incorrect co-translational ubiquitination and degradation [80]. Pulse-labeling experiments revealed an increased ubiquitination of nascent chains upon loss of NAC. Thus, NAC protects nascent unfolded polypeptides as they emerge from the ribosome. Many nascent chains shielded by NAC are quite long, suggesting that NAC does not only function by sterically blocking the access of the ubiquitin-proteasome system to the ribosome [80] .
4 The Ribosome-Associated Hsp70/Hsp40 System
Besides NAC a second ribosome-associated chaperone system consisting of Hsp70 and Hsp40 chaperones is found in association with eukaryotic ribosomes (Figs. 2.1c and 2.2a). The RAC is the central component of this system and conserved from yeast to mammals [4] (Fig. 2.1c). RAC is a stable heterodimer formed by an Hsp40 chaperone called Zuotin (Zuo) and an Hsp70 called Ssz [10, 11, 81] . In S. cerevisiae this system is extended by another ribosome-anchored chaperone, the Hsp70 Ssb (Fig. 2.1c) . Ssb binds to the nascent polypeptide, which requires the activity of RAC [11, 82] (Fig. 2.2a). RAC stimulates the ATPase activity of Ssb and thus enhances its affinity for unfolded polypeptides, but RAC seems not to bind to substrate proteins itself (Fig. 2.2a) [83]. Within the RAC complex, Zuo contacts Ssb by its J-domain and in addition anchors RAC at the ribosome [84–86] (Fig. 2.1c). The function of Ssz is less clear. It might fulfill regulatory functions by inducing structural rearrangements within the J-domain of Zuo, which in turn might strengthen the contact to Ssb [87]. Alternatively, Ssz might play a role in the recruitment of substrates to Ssb [84]. However, no experimental data exist so far to support this hypothesis.
In the yeast genome, two genes (SSB1 and SSB2) encode the proteins Ssb1 and Ssb2 that differ only in four amino acids. Therefore, in the following, they are collectively referred to as Ssb. The functional cooperation between the members of the yeast Hsp70/Hsp40-chaperone triad was discovered by genetic experiments. Yeast cells lacking either one or all three proteins of this system show similar phenotypes: sensitivity to high salt concentrations, hypersensitivity to aminoglycosides that increased translational error rates, and cold-sensitivity [11, 82, 86, 88, 89]. The first hint that the RAC-Ssb system might be involved in the folding of nascent chains came from cross-linking experiments showing that Ssb is able to contact short nascent polypeptide chains and this interaction was dependent on RAC [11, 82, 90]. This suggests that Ssb acts as a chaperone for nascent polypeptides. In addition, these data indicate that the chaperone triad must bind to ribosomes in close proximity to the ribosomal tunnel exit site. Moreover, the finding that expression of the prokaryotic ribosome-associated chaperone TF could partially alleviate the aminoglycoside sensitivity of triad-deficient yeast cells indicates overlapping functions of the two chaperone systems from different kingdoms of life [91]. A more recent study shows that the RAC-Ssb and the NAC systems are functionally connected in yeast [77]. The simultaneous deletion of NAC- and Ssb-encoding genes caused conditional loss of cell viability under protein-folding stress conditions. Furthermore, loss of Ssb resulted in the aggregation of newly synthesized polypeptides, ribosomal proteins, as well as several ribosome biogenesis factors. Likewise, the levels of translating ribosomes and 60S ribosomal subunits were decreased in ssb∆ cells. These defects were aggravated when NAC was absent in addition to the Ssb chaperone. These findings suggest that not only the folding of cytosolic proteins is affected by these chaperones but also ribosome biogenesis.
Based on the finding that mainly ribosomal proteins are aggregation-prone in the absence of Ssb and on a recent study investigating the nascent interactome of Ssb, it is likely that ribosomal proteins represent important substrates of this chaperone [77, 92].
Ssb represents a member of the Hsp70 chaperone family with a N-terminal nucleotide-binding domain (NBD) and a C-terminal substrate-binding domain (SBD)(Fig. 2.1c). Additionally, it contains a potential nuclear export sequence (NES) at its C-terminus (Fig. 2.1c) [93]. Ssb is found primarily in the cytosol, upon mutation of the NES however, it strongly accumulates in the nucleus [93]. This suggests that Ssb may rapidly shuttle between the nucleus and cytosol.
Like Ssb, also the Hsp70 Ssz is composed of a NBD and SBD . In contrast to other Hsp70 chaperones, Ssz is not able to hydrolyze ATP and up to now no binding of Ssz to a substrate could be detected [81, 83]. One reason might be its shorter substrate-binding domain (Fig. 2.1c) [90]. Furthermore, it was suggested that amino acid substitutions at three positions in the ATP-binding pocket (compared to other Hsp70s) might be the reason for the inability of Ssz to hydrolyze ATP [84]. As Ssz is continuously in an ATP-bound state its affinity for potential substrates is assumed to be rather low. It was proposed that Ssz could potentially act as a low-affinity holding chaperone that guides the growing nascent chain from the tunnel to Ssb; an interesting aspect that needs to be experimentally tested in the future [84].
The other subunit of RAC, Zuo, consists of an unstructured N-terminal domain that is followed by a J-domain. The J-domain has a conserved HPD (Histidine, Proline, Aspartic acid) motif and is required to stimulate the ATPase activity of Ssb. A characteristic feature of Zuo is a highly positively charged region (charged region) that is located near its C-terminus (Fig. 2.1c) [10, 86, 94]. This region was discussed for a long time to be involved in the ribosome association of Zuo [85, 86].
It was shown that the highly flexible and unstructured N-terminus of Zuo is responsible for the formation of the RAC complex . The first 62 N-terminal amino acids of Zuo contact both domains of Ssz. This interaction is crucial and results in the formation of the unusual, stable chaperone-chaperone complex RAC [87]. Interestingly, upon complex formation the J-domain of Zuo becomes more dynamic. As the J-domain is required for the stimulation of the ATPase activity of Ssb, the complex formation with Ssz might induce a conformation of Zuo that favors the interaction with Ssb and thus might be crucial for the function of RAC as a co-chaperone [87].
Recently, the crystal structure of a truncated RAC version from the thermophilic fungus Chaetomium thermophilium could be solved [84]. The structure was modeled into small-angle X-ray scattering (SAXS) densities of RAC bound to the ribosome. The data revealed that RAC forms an elongated complex of 180 Å in length (Fig. 2.1c) that binds to the ribosome near the ribosomal proteins L22 and L31 (Fig. 2.1c). In combination with cryo-electron microscopy data of ribosome-bound RAC, it was shown that the complex bends over the ribosomal tunnel exit site and contacts the ribosomal extension segment ES27 that stabilizes a distinct conformation of RAC. The elongated RAC structure can be divided into a head, neck and a body region (Fig. 2.1c). Whereas the large body is mainly composed of Ssz, the head is formed by Zuo and mediates the ribosome association (Fig. 2.1c). In agreement with previous data, it could be shown that ribosome association is mediated by a helix that is formed by the C-terminal part of the charged region [84].
While Ssb is found exclusively in fungi, homologs of RAC were identified in mammals as well [95–97] (Fig. 2.1c). This indicates that the presence of Hsp70/40 chaperones on ribosomes is common in the eukaryotic world (Figs. 2.1c and 2.2a). Supporting this assumption, the human homologs of Ssz (Hsp70L1) and Zuo (MPP11) can partially substitute for the loss of RAC in yeast [95–97]. Additionally, the knockdown of human MPP11 in HeLa cells results in growth defects and sensitivity towards drugs similar to what was observed for yeast cells lacking RAC [96]. These findings indicate that RAC fulfills similar functions in yeast as well as in metazoans. Despite these similarities between yeast and mammalian RAC, there are also large structural and functional differences. Most strikingly, complementation of yeast RAC with the mammalian system is independent of Ssb [97]. It was thus suggested recently that cytosolic Hsp70s, which cannot bind to the ribosome on their own, act as functional partners for RAC in higher eukaryotes (Fig. 2.2a) [96]. In addition, MPP11 has some variation in its domain composition compared to Zuo including two additional SANT-like domains at the C-terminal end of the chaperone (Fig. 2.1c). SANT (Swi3, Ada2, N-Cor, TfIIIB) domains are normally involved in DNA binding and chromatin remodeling, however, their role in MPP11 is unknown.
A recent analysis directly addressed the substrate-specificity of ribosome-bound Ssb [92]. In an elegant approach, a TAP-tagged Ssb version was expressed in yeast and used to pull down associated ribosomes along with the translated mRNA. Subsequently, the mRNAs were subjected to sequence analysis to determine the co-translational interactome of Ssb. The data revealed that Ssb preferentially binds to nascent cytosolic and nuclear proteins, but not secretory polypeptides, which are targeted by SRP (Fig. 2.2b). Moreover, about 80 % of all nascent cytosolic and nuclear proteins interact with Ssb. A common feature among the Ssb substrates is that they are large in size, have a low α-helical content, and an increased content of hydrophobic elements. Additionally, Ssb interacts with many nascent subunits of oligomeric complexes and proteins that are engaged in a large number of interactions (Fig. 2.2b), for example, all subunits of the chaperonin complex TRiC, most subunits of the proteasome, and a large set of ribosomal proteins associated with Ssb during their synthesis [92]. In contrast, the abundance of a protein had no influence on Ssb binding. Thus, Ssb may assist co-translational folding of large proteins with complicated structures or that require binding partners for their stability. This also agrees with a potential role for Ssb in stabilizing free subunits of oligomeric complexes, which may expose contact sites that are prone to undergo false interactions. In a next step, the authors of the same study analyzed if RAC modulates the substrate specificity of Ssb [92]. To this end, they compared the substrate interactions of Ssb in a wild type yeast strain and in a strain lacking both RAC subunits. They indeed observed an influence of RAC on substrate binding of Ssb. While a subset of Ssb-substrate interactions was unaffected by the loss of RAC, others were significantly altered. For example, the interaction between Ssb and nascent cytosolic proteins was reduced while the interaction with membrane and mitochondrial proteins was enhanced in cells lacking RAC. Thus, RAC influences the co-translational substrate-specificity of Ssb [92].
5 Versatility of Ribosome-Associated Chaperones and their off Ribosomal Functions
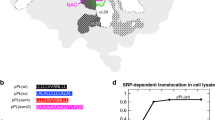
The function of ribosome-associated chaperones was initially thought to be limited to assistance in de novo folding of newly synthesized proteins [5]. However, all ribosome-associated chaperones known so far bind to the ribosome only transiently and thus are additionally found in a non-ribosomal state in the cytosol. There is increasing evidence that ribosome-associated chaperones are not in an inactive state when they are off the ribosome rather they are assumed to display additional functions (Fig. 2.3). In a ribosome-associated state, they all contact the nascent chain and support the de novo folding of newly synthesized proteins by different modes of action. In contrast, the functional spectrum in a non-ribosome-associated state is much more diverse and less well understood.
a When bound to the ribosome trigger factor (TF) protects cytosolic nascent polypeptides (yellow) from aggregation, proteolysis, and premature folding. Thus, TF supports correct de novo protein folding . Thereby, especially outer membrane proteins strongly bind to TF, and TF was shown to facilitate their correct targeting and assembly. In addition, TF is suggested to facilitate the assembly of large protein oligomers and assemblies independent from its function at the ribosome. b In yeast, nascent polypeptide-associated complex (NAC) and ribosome-associated complex (RAC)–Ssb bind to ribosomes and interact with nascent polypeptide chains to support de novo folding. NAC is suggested to modulate SRP-dependent cotranslational targeting of secretory proteins to the endoplasmic reticulum (ER) translocon. Moreover, NAC interacts with various types of protein aggregates and supports the resolubilization of heat-shock-induced protein aggregates. Non-ribosomal Ssb seems to be involved in glucose signaling by regulating the phosphorylation status of Snf1. Ssb-RAC and NAC have also additional functions in the nucleus. Ssb-RAC and NAC are suggested to be involved in ribosome biogenesis and Zuo or the entire RAC was suggested to interact with the transcription factor Pdr1, which initiates the pleiotropic drug resistance (PDR)
E. coli TF was found to co-purify with several full-length proteins including many ribosomal proteins, such as ribosomal protein S7 [19]. Interestingly, many of the interacting proteins were previously found to be aggregation-prone in E. coli cells lacking TF and the cytosolic Hsp70 chaperone DnaK [32]. To further investigate the non-ribosomal interaction of TF with substrates, Hendrickson and co-workers co-crystallized TF with the S7 protein, both derived from.Thermotoga maritima [19]. The structural data revealed that TF binds to almost natively folded S7 proteins in a 2:2 stoichiometry. Through the binding of the TF molecules, large surfaces of the S7 are masked which are normally buried in the interior of the 30S ribosomal subunit. Accordingly, TF might not only co-translationally support protein folding but also function post-translationally in the biogenesis of large macromolecular assemblies such as ribosomes by stabilizing newly synthesized proteins until they become incorporated into complexes [19] (Fig. 2.3a)
Also eukaryotic ribosome-tethered chaperones display off-ribosomal functions. It was shown earlier that depending on the characteristics of the nascent chain between 17.8 and 41.8 % of the Ssb molecules, 12.3–16.2 % of the RAC molecules, and 29.3–66.4 % of all NAC molecules are found in association with actively translating ribosomes in an in vitro translation extract derived from yeast cells while the other molecules diffuse freely [41]. Cells lacking Ssb accumulate insoluble newly synthesized proteins and additionally reveal a strong reduction in translation along with a strong decrease in the levels of ribosomal particles, in particular of the large 60S ribosomal subunit. In line with this finding, Ssb as well as Zuo show genetic interactions with Jjj1, a chaperone specific for ribosome biogenesis [77, 98]. Moreover, Zuo associates with nuclear 60S ribosomal biogenesis intermediates and Zuo (together with Ssb) also participates in the maturation of the 35S ribosomal RNA (rRNA) [98]. Thus, the RAC-Ssb system seems to play an import role in the biogenesis of ribosomal 60S particles (Fig. 2.3b). Whether Ssb and Zuo act independently of each other in ribosome biogenesis or—similar to their activity on ribosomes—always as a RAC-Ssb system is unclear (Fig. 2.3b).
Ribosome biogenesis is a highly complex and energy-consuming process and thus has to be tightly regulated. In eukaryotes, the ribosomal proteins are synthesized in the cytosol and subsequently imported into the nucleus where the assembly of the pre-ribosomal particle takes place. With the help of a variety of ribosome biogenesis factors, the pre-ribosomes undergo several processing steps and are transported out of the nucleus into the cytosol [99]. How might RAC-Ssb act during the biogenesis of new ribosomal particles? One possibility is that the RAC-Ssb system is required for the co-translational folding and prevention of aggregation of ribosomal proteins and, likewise, Ssb and/or Zuo may additionally assist the transport for the newly synthesized ribosomal proteins into the nucleus where the ribosomal proteins become incorporated into pre-ribosomal particles. In this scenario, Ssb and/or Zuo would stay bound to newly synthesized ribosomal proteins even after their release from the ribosome and accompany them during the transport to the nucleus. Thus, the RAC-Ssb system could act by preventing the aggregation of ribosomal proteins after their synthesis and before they become stabilized by incorporation into a new ribosomal subunit. Alternatively and not mutually exclusive to this hypothesis, Ssb and Zuo could act by directly promoting ribosome biogenesis in the nucleus, for example, by supporting the processing of rRNA or by modulating the activity of certain ribosome biogenesis factors. The finding that Zuo and Ssb are also detectable in the nucleus albeit only very transiently supports the idea that RAC-Ssb is not only engaged in the co-translational folding of ribosomal proteins in the cytosol but also fulfills functions in the nucleus besides its action on the ribosome [93, 98] (Fig. 2.3b).
Additionally, it was suggested that Ssb regulates the phosphorylation state of the kinase Snf1 and that this function is independent of its ribosome association [100] (Fig. 2.3b). Snf1 is active in a phosphorylated state under non-glucose conditions and drives the expression of glucose-repressed genes. Ssb, however, was shown to be required to keep Snf1 unphosphorylated and inactive when glucose is available. Thus, Ssb might play a role in the appropriate response to changing glucose concentrations [100].
Both RAC subunits where shown to be able to activate the transcription factor Pdr1 resulting in the induction of the pleiotropic drug resistance (PDR) [101–103] (Fig. 2.3b). The PDR is a highly specific transcriptional response that leads to the upregulation of a set of genes involved in the resistance towards several drugs and toxic agents in the cellular environment. For example, Pdr1 target genes include several ATP-binding cassette transporters that extrude xenobiotics from cells, rendering them resistant to a variety of toxic compounds [101]. For activation of Pdr1 in S. cerevisae, the ATPase domain of Ssz or the 13C-terminal amino acids of Zuo are required and sufficient. Interestingly, when Zuo is bound to the ribosome, its C-terminal 86 residues form a left-handed four helix bundle that buries the residues critical for activation of Pdr1 within its interior. However, the simple dissociation of Zuo1 from the ribosome is not sufficient for Pdr1 transcriptional activity but requires the additional unfolding of the C-terminus of Zuo. How this unfolding is regulated is not clear so far [101]. For Ssz too, it was shown that it has to be free of ribosomes to induce the PDR [101, 103]. These findings imply that the role of the RAC components in the PDR it distinct from their roles as ribosome-associated chaperones (Fig. 2.3b).
A very recent study unraveled a new function of NAC during misfolding and aggregation of cytosolic and nuclear proteins beyond its activity on ribosomes. It was shown that C. elegans NAC interacts with different types of amorphous aggregates, like aggregates that accumulate during aging or upon heat shock, as well as with amyloid-like Aβ fibrils [78] (Fig. 2.3b). These findings are in agreement with another study that identified human NAC to associate with artificial β-sheet fibrils [104]. The physical interaction between NAC and aggregates has also physiological consequences, as disaggregation of heat shock protein aggregates in C.elegans is dependent on the presence of NAC [78] (Fig. 2.3b). However, it is not clear how NAC is able to support disaggregation in metazoans. As NAC has no ATPase activity and thus cannot apply mechanical force, its mechanism must be different from the one of classical disaggregases such as the Hsp70-Hsp40-Hsp100 system [105]. One possibility is that NAC in C. elegans interacts with non-native proteins during the aggregation process and thereby becomes itself associated with aggregates. NAC associated with aggregates perhaps recruits other chaperones with the capacity to remodel the aggregates, e.g., Hsp70/40 and Hsp110 type chaperones. This is in line with the finding that NAC co-immunoprecipitates with many other chaperones [78]. Thus, NAC is able to dissociate from the ribosome and to re-localize towards misfolded and insoluble proteins. Importantly, the depletion of NAC by RNA interference (RNAi) as well as the re-localization of NAC to protein aggregates upon protein stress conditions is associated with a strong decline of translation in the cytosol in C. elegans. This suggests that NAC is also able to modulate ribosome activity. Thus, NAC has a housekeeping function on ribosomes to promote folding and translation while under proteotoxic stress , its action is shifted towards aggregated proteins. This has beneficial consequences: NAC can aid in the re-solubilization of misfolded proteins in cooperation with other molecular chaperones and, likewise, translation and thus the influx of new proteins that need to be assisted by chaperones in their folding program is reduced. The re-localization is reversible, as NAC can re-associate with the ribosome once misfolded and aggregated proteins are eliminated and proteostasis is re-balanced. Based on these observations, it was suggested that the functional depletion of NAC from ribosomes into aggregates is an important regulatory circuit to modulate translation [78, 106].
In sum, there is strong evidence that ribosome-associated chaperones are versatile and vital elements of the chaperone network that interact in vivo with a large variety of substrates to perform more than one function (Fig. 2.3). They co-translationally support de novo folding and also act by unknown mechanisms to regulate ribosome biogenesis and translation, which regulates the influx of new proteins into the cellular proteome. In addition, there is a stress-related function at least for NAC. This complex associates with protein aggregates and, as a consequence, either directly or indirectly dims protein synthesis to reduce the new protein load for chaperones and to allow the cells to recover from proteotoxic stress . It will be exciting to illuminate the individual roles, mechanisms, and substrate specificities of these chaperones in their functions on and off the ribosome.
References
Hartl FU, Hayer-Hartl M (2009) Converging concepts of protein folding in vitro and in vivo. Nat Struct Mol Biol 16(6):574–581. doi:10.1038/nsmb.1591
Hartl FU, Bracher A, Hayer-Hartl M (2011) Molecular chaperones in protein folding and proteostasis. Nature 475(7356):324–332. doi:10.1038/nature10317
Bukau B, Deuerling E, Pfund C, Craig EA (2000) Getting newly synthesized proteins into shape. Cell 101(2):119–122. doi:10.1016/S0092-8674(00)80806-5
Preissler S, Deuerling E (2012) Ribosome-associated chaperones as key players in proteostasis. Trends Biochem Sci 37(7):274–283. doi:10.1016/j.tibs.2012.03.002
Hartl FU, Hayer-Hartl M (2002) Molecular chaperones in the cytosol: from nascent chain to folded protein. Science 295(5561):1852–1858. doi:10.1126/science.1068408
Maier T, Ferbitz L, Deuerling E, Ban N (2005) A cradle for new proteins: trigger factor at the ribosome. Curr Opin Struct Biol 15(2):204–212. doi:10.1016/j.sbi.2005.03.005
Hoffmann A, Bukau B, Kramer G (2010) Structure and function of the molecular chaperone trigger factor. Biochim Biophys Acta 1803(6):650–661. doi:10.1016/j.bbamcr.2010.01.017
Hoffmann A, Becker AH, Zachmann-Brand B, Deuerling E, Bukau B, Kramer G (2012) Concerted action of the ribosome and the associated chaperone trigger factor confines nascent polypeptide folding. Mol Cell 48(1):63–74. doi:10.1016/j.molcel.2012.07.018
Ferbitz L, Maier T, Patzelt H, Bukau B, Deuerling E, Ban N (2004) Trigger factor in complex with the ribosome forms a molecular cradle for nascent proteins. Nature 431(7008):590–596. doi:10.1038/nature02899
Gautschi M, Lilie H, Funfschilling U, Mun A, Ross S, Lithgow T, Rucknagel P, Rospert S (2001) RAC, a stable ribosome-associated complex in yeast formed by the DnaK-DnaJ homologs Ssz1p and Zuo. Proc Natl Acad Sci U S A 98(7):3762–3767. doi:10.1073/pnas.071057198
Gautschi M, Mun A, Ross S, Rospert S (2002) A functional chaperone triad on the yeast ribosome. Proc Natl Acad Sci U S A 99(7):4209–4214. doi:10.1073/pnas.062048599
Stoller G, Rucknagel KP, Nierhaus KH, Schmid FX, Fischer G, Rahfeld JU (1995) A ribosome-associated peptidyl-prolyl cis/trans isomerase identified as the trigger factor. EMBO J 14(20):4939–4948
Kramer G, Rauch T, Rist W, Vorderwulbecke S, Patzelt H, Schulze-Specking A, Ban N, Deuerling E, Bukau B (2002) L23 protein functions as a chaperone docking site on the ribosome. Nature 419(6903):171–174. doi:10.1038/nature01047
Stoller G, Tradler T, Rucknagel KP, Rahfeld JU, Fischer G (1996) An 11.8 kDa proteolytic fragment of the E. coli trigger factor represents the domain carrying the peptidyl-prolyl cis/trans isomerase activity. FEBS Lett 384(2):117–122
Gupta R, Lakshmipathy SK, Chang HC, Etchells SA, Hartl FU (2010) Trigger factor lacking the PPIase domain can enhance the folding of eukaryotic multi-domain proteins in Escherichia coli. FEBS Lett 584(16):3620–3624. doi:10.1016/j.febslet.2010.07.036
Liu CP, Zhou QM, Fan DJ, Zhou JM (2010) PPIase domain of trigger factor acts as auxiliary chaperone site to assist the folding of protein substrates bound to the crevice of trigger factor. Int J Biochem Cell Biol 42(6):890–901. doi:10.1016/j.biocel.2010.01.019
Kramer G, Patzelt H, Rauch T, Kurz TA, Vorderwulbecke S, Bukau B, Deuerling E (2004) Trigger factor peptidyl-prolyl cis/trans isomerase activity is not essential for the folding of cytosolic proteins in Escherichia coli. J Biol Chem 279(14):14165–14170. doi:10.1074/jbc.M313635200
Merz F, Boehringer D, Schaffitzel C, Preissler S, Hoffmann A, Maier T, Rutkowska A, Lozza J, Ban N, Bukau B, Deuerling E (2008) Molecular mechanism and structure of Trigger Factor bound to the translating ribosome. Embo J 27(11):1622–1632. doi:10.1038/emboj.2008.89
Martinez-Hackert E, Hendrickson WA (2009) Promiscuous substrate recognition in folding and assembly activities of the trigger factor chaperone. Cell 138(5):923–934. doi:10.1016/j.cell.2009.07.044
Rutkowska A, Mayer MP, Hoffmann A, Merz F, Zachmann-Brand B, Schaffitzel C, Ban N, Deuerling E, Bukau B (2008) Dynamics of trigger factor interaction with translating ribosomes. J Biol Chem 283(7):4124–4132. doi:10.1074/jbc.M708294200
Hesterkamp T, Deuerling E, Bukau B (1997) The amino-terminal 118 amino acids of Escherichia coli trigger factor constitute a domain that is necessary and sufficient for binding to ribosomes. J Biol Chem 272(35):21865–21871
Patzelt H, Kramer G, Rauch T, Schonfeld HJ, Bukau B, Deuerling E (2002) Three-state equilibrium of Escherichia coli trigger factor. Biol Chem 383(10):1611–1619. doi:10.1515/BC.2002.182
Zarnt T, Tradler T, Stoller G, Scholz C, Schmid FX, Fischer G (1997) Modular structure of the trigger factor required for high activity in protein folding. J Mol Biol 271(5):827–837. doi:10.1006/jmbi.1997.1206
Raine A, Lovmar M, Wikberg J, Ehrenberg M (2006) Trigger factor binding to ribosomes with nascent peptide chains of varying lengths and sequences. J Biol Chem 281(38):28033–28038. doi:10.1074/jbc.M605753200
Maier R, Eckert B, Scholz C, Lilie H, Schmid FX (2003) Interaction of trigger factor with the ribosome. J Mol Biol 326(2):585–592
Kaiser CM, Chang HC, Agashe VR, Lakshmipathy SK, Etchells SA, Hayer-Hartl M, Hartl FU, Barral JM (2006) Real-time observation of trigger factor function on translating ribosomes. Nature 444(7118):455–460. doi:10.1038/nature05225
Oh E, Becker AH, Sandikci A, Huber D, Chaba R, Gloge F, Nichols RJ, Typas A, Gross CA, Kramer G, Weissman JS, Bukau B (2011) Selective ribosome profiling reveals the cotranslational chaperone action of trigger factor in vivo. Cell 147(6):1295–1308. doi:10.1016/j.cell.2011.10.044
Kramer G, Boehringer D, Ban N, Bukau B (2009) The ribosome as a platform for co-translational processing, folding and targeting of newly synthesized proteins. Nat Struct Mol Biol 16(6):589–597. doi:10.1038/nsmb.1614
Sandikci A, Gloge F, Martinez M, Mayer MP, Wade R, Bukau B, Kramer G (2013) Dynamic enzyme docking to the ribosome coordinates N-terminal processing with polypeptide folding. Nat Struct Mol Biol 20(7):843–850. doi:10.1038/nsmb.2615
Lakshmipathy SK, Gupta R, Pinkert S, Etchells SA, Hartl FU (2010) Versatility of trigger factor interactions with ribosome-nascent chain complexes. J Biol Chem 285(36):27911–27923. doi:10.1074/jbc.M110.134163
Crooke E, Wickner W (1987) Trigger factor: a soluble protein that folds pro-OmpA into a membrane-assembly-competent form. Proc Natl Acad Sci U S A 84(15):5216–5220
Deuerling E, Schulze-Specking A, Tomoyasu T, Mogk A, Bukau B (1999) Trigger factor and DnaK cooperate in folding of newly synthesized proteins. Nature 400(6745):693–696. doi:10.1038/23301
Deuerling E, Patzelt H, Vorderwulbecke S, Rauch T, Kramer G, Schaffitzel E, Mogk A, Schulze-Specking A, Langen H, Bukau B (2003) Trigger factor and DnaK possess overlapping substrate pools and binding specificities. Mol Microbiol 47(5):1317–1328
Teter SA, Houry WA, Ang D, Tradler T, Rockabrand D, Fischer G, Blum P, Georgopoulos C, Hartl FU (1999) Polypeptide flux through bacterial Hsp70: DnaK cooperates with trigger factor in chaperoning nascent chains. Cell 97(6):755–765
Agashe VR, Guha S, Chang HC, Genevaux P, Hayer-Hartl M, Stemp M, Georgopoulos C, Hartl FU, Barral JM (2004) Function of trigger factor and DnaK in multidomain protein folding: increase in yield at the expense of folding speed. Cell 117(2):199–209
Patzelt H, Rudiger S, Brehmer D, Kramer G, Vorderwulbecke S, Schaffitzel E, Waitz A, Hesterkamp T, Dong L, Schneider-Mergener J, Bukau B, Deuerling E (2001) Binding specificity of Escherichia coli trigger factor. Proc Natl Acad Sci U S A 98(25):14244–14249. doi:10.1073/pnas.261432298
Hoffmann A, Merz F, Rutkowska A, Zachmann-Brand B, Deuerling E, Bukau B (2006) Trigger factor forms a protective shield for nascent polypeptides at the ribosome. J Biol Chem 281(10):6539–6545. doi:10.1074/jbc.M512345200
Tomic S, Johnson AE, Hartl FU, Etchells SA (2006) Exploring the capacity of trigger factor to function as a shield for ribosome bound polypeptide chains. FEBS Lett 580(1):72–76. doi:10.1016/j.febslet.2005.11.050
Mashaghi A, Kramer G, Bechtluft P, Zachmann-Brand B, Driessen AJ, Bukau B, Tans SJ (2013) Reshaping of the conformational search of a protein by the chaperone trigger factor. Nature 500(7460):98–101. doi:10.1038/nature12293
Becker AH, Oh E, Weissman JS, Kramer G, Bukau B (2013) Selective ribosome profiling as a tool for studying the interaction of chaperones and targeting factors with nascent polypeptide chains and ribosomes. Nat Protoc 8(11):2212–2239. doi:10.1038/nprot.2013.133
Raue U, Oellerer S, Rospert S (2007) Association of protein biogenesis factors at the yeast ribosomal tunnel exit is affected by the translational status and nascent polypeptide sequence. J Biol Chem 282(11):7809–7816. doi:10.1074/jbc.M611436200
Wiedmann B, Sakai H, Davis TA, Wiedmann M (1994) A protein complex required for signal-sequence-specific sorting and translocation. Nature 370(6489):434–440. doi:10.1038/370434a0
Beatrix B, Sakai H, Wiedmann M (2000) The alpha and beta subunit of the nascent polypeptide-associated complex have distinct functions. J Biol Chem 275(48):37838–37845. doi:10.1074/jbc.M006368200
Reimann B, Bradsher J, Franke J, Hartmann E, Wiedmann M, Prehn S, Wiedmann B (1999) Initial characterization of the nascent polypeptide-associated complex in yeast. Yeast 15(5):397–407. doi:10.1002/(SICI)1097-0061(19990330)15:5<397::AID-YEA384>3.0.CO;2-U
Wang S, Sakai H, Wiedmann M (1995) NAC covers ribosome-associated nascent chains thereby forming a protective environment for regions of nascent chains just emerging from the peptidyl transferase center. J Cell Biol 130(3):519–528
Lauring B, Wang S, Sakai H, Davis TA, Wiedmann B, Kreibich G, Wiedmann M (1995) Nascent-polypeptide-associated complex: a bridge between ribosome and cytosol. Cold Spring Harb Symp Quant Biol 60:47–56
Rospert S, Dubaquie Y, Gautschi M (2002) Nascent-polypeptide-associated complex. Cell Mol Life Sci 59(10):1632–1639
Liu Y, Hu Y, Li X, Niu L, Teng M (2010) The crystal structure of the human nascent polypeptide-associated complex domain reveals a nucleic acid-binding region on the NACA subunit. BioChemistry 49(13):2890–2896. doi:10.1021/bi902050p
Wang L, Zhang W, Zhang XC, Li X, Rao Z (2010) Crystal structures of NAC domains of human nascent polypeptide-associated complex (NAC) and its alphaNAC subunit. Protein Cell 1(4):406–416. doi:10.1007/s13238-010-0049-3
Spreter T, Pech M, Beatrix B (2005) The crystal structure of archaeal nascent polypeptide-associated complex (NAC) reveals a unique fold and the presence of a ubiquitin-associated domain. J Biol Chem 280(16):15849–15854. doi:10.1074/jbc.M500160200
Hurley JH, Lee S, Prag G (2006) Ubiquitin-binding domains. Biochem J 399(3):361–372. doi:10.1042/BJ20061138
Madura K (2002) The ubiquitin-associated (UBA) domain: on the path from prudence to prurience. Cell Cycle 1(4):235–244
Searle MS, Garner TP, Strachan J, Long J, Adlington J, Cavey JR, Shaw B, Layfield R (2012) Structural insights into specificity and diversity in mechanisms of ubiquitin recognition by ubiquitin-binding domains. Biochem Soc Trans 40(2):404–408. doi:10.1042/BST20110729
Su V, Lau AF (2009) Ubiquitin-like and ubiquitin-associated domain proteins: significance in proteasomal degradation. Cell Mol Life Sci 66(17):2819–2833. doi:10.1007/s00018-009-0048-9
Wegrzyn RD, Hofmann D, Merz F, Nikolay R, Rauch T, Graf C, Deuerling E (2006) A conserved motif is prerequisite for the interaction of NAC with ribosomal protein L23 and nascent chains. J Biol Chem 281(5):2847–2857. doi:10.1074/jbc.M511420200
Pech M, Spreter T, Beckmann R, Beatrix B (2010) Dual binding mode of the nascent polypeptide-associated complex reveals a novel universal adapter site on the ribosome. J Biol Chem 285(25):19679–19687. doi:10.1074/jbc.M109.092536
Zhang Y, Berndt U, Golz H, Tais A, Oellerer S, Wolfle T, Fitzke E, Rospert S (2012) NAC functions as a modulator of SRP during the early steps of protein targeting to the endoplasmic reticulum. Mol Biol Cell 23(16):3027–3040. doi:10.1091/mbc.E12-02-0112
Lauring B, Kreibich G, Weidmann M (1995) The intrinsic ability of ribosomes to bind to endoplasmic reticulum membranes is regulated by signal recognition particle and nascent-polypeptide-associated complex. Proc Natl Acad Sci U S A 92(21):9435–9439
Lauring B, Sakai H, Kreibich G, Wiedmann M (1995) Nascent polypeptide-associated complex protein prevents mistargeting of nascent chains to the endoplasmic reticulum. Proc Natl Acad Sci U S A 92(12):5411–5415
Moller I, Beatrix B, Kreibich G, Sakai H, Lauring B, Wiedmann M (1998) Unregulated exposure of the ribosomal M-site caused by NAC depletion results in delivery of non-secretory polypeptides to the Sec61 complex. FEBS Lett 441(1):1–5
Moller I, Jung M, Beatrix B, Levy R, Kreibich G, Zimmermann R, Wiedmann M, Lauring B (1998) A general mechanism for regulation of access to the translocon: competition for a membrane attachment site on ribosomes. Proc Natl Acad Sci U S A 95(23):13425–13430
Powers T, Walter P (1996) The nascent polypeptide-associated complex modulates interactions between the signal recognition particle and the ribosome. Curr Biol 6(3):331–338
Raden D, Gilmore R (1998) Signal recognition particle-dependent targeting of ribosomes to the rough endoplasmic reticulum in the absence and presence of the nascent polypeptide-associated complex. Mol Biol Cell 9(1):117–130
Wickner W (1995) The nascent-polypeptide-associated complex: having a “NAC” for fidelity in translocation. Proc Natl Acad Sci U S A 92(21):9433–9434
Kim SH, Shim KS, Lubec G (2002) Human brain nascent polypeptide-associated complex alpha subunit is decreased in patients with Alzheimer’ s disease and Down syndrome. J Investig Med 50(4):293–301
Thiede B, Dimmler C, Siejak F, Rudel T (2001) Predominant identification of RNA-binding proteins in Fas-induced apoptosis by proteome analysis. J Biol Chem 276(28):26044–26050. doi:10.1074/jbc.M101062200
Bloss TA, Witze ES, Rothman JH (2003) Suppression of CED-3-independent apoptosis by mitochondrial betaNAC in Caenorhabditis elegans. Nature 424(6952):1066–1071. doi:10.1038/nature01920
Quelo I, Hurtubise M, St-Arnaud R (2002) alphaNAC requires an interaction with c-Jun to exert its transcriptional coactivation. Gene Expr 10(5-6):255–262
Yotov WV, Moreau A, St-Arnaud R (1998) The alpha chain of the nascent polypeptide-associated complex functions as a transcriptional coactivator. Mol Cell Biol 18(3):1303–1311
Yotov WV, St-Arnaud R (1996) Differential splicing-in of a proline-rich exon converts alphaNAC into a muscle-specific transcription factor. Genes Dev 10(14):1763–1772
Moreau A, Yotov WV, Glorieux FH, St-Arnaud R (1998) Bone-specific expression of the alpha chain of the nascent polypeptide-associated complex, a coactivator potentiating c-Jun-mediated transcription. Mol Cell Biol 18(3):1312–1321
Markesich DC, Gajewski KM, Nazimiec ME, Beckingham K (2000) bicaudal encodes the Drosophila beta NAC homolog, a component of the ribosomal translational machinery*. Development 127(3):559–572
Deng JM, Behringer RR (1995) An insertional mutation in the BTF3 transcription factor gene leads to an early postimplantation lethality in mice. Transgenic Res 4(4):264–269
Deuerling E, Bukau B (2004) Chaperone-assisted folding of newly synthesized proteins in the cytosol. Crit Rev Biochem Mol Biol 39(5-6):261–277. doi:10.1080/10409230490892496
Pechmann S, Willmund F, Frydman J (2013) The ribosome as a hub for protein quality control. Mol Cell 49(3):411–421. doi:10.1016/j.molcel.2013.01.020
Wegrzyn RD, Deuerling E (2005) Molecular guardians for newborn proteins: ribosome-associated chaperones and their role in protein folding. Cell Mol Life Sci 62(23):2727–2738. doi:10.1007/s00018-005-5292-z
Koplin A, Preissler S, Ilina Y, Koch M, Scior A, Erhardt M, Deuerling E (2010) A dual function for chaperones SSB-RAC and the NAC nascent polypeptide-associated complex on ribosomes. J Cell Biol 189(1):57–68. doi:10.1083/jcb.200910074
Kirstein-Miles J, Scior A, Deuerling E, Morimoto RI (2013) The nascent polypeptide-associated complex is a key regulator of proteostasis. EMBO J 32(10):1451–1468. doi:10.1038/emboj.2013.87
del Alamo M, Hogan DJ, Pechmann S, Albanese V, Brown PO, Frydman J (2011) Defining the specificity of cotranslationally acting chaperones by systematic analysis of mRNAs associated with ribosome-nascent chain complexes. PLoS Biol 9(7):e1001100. doi:10.1371/journal.pbio.1001100
Duttler S, Pechmann S, Frydman J (2013) Principles of cotranslational ubiquitination and quality control at the ribosome. Mol Cell 50(3):379–393. doi:10.1016/j.molcel.2013.03.010
Huang P, Gautschi M, Walter W, Rospert S, Craig EA (2005) The Hsp70 Ssz1 modulates the function of the ribosome-associated J-protein Zuo1. Nat Struct Mol Biol 12(6):497–504. doi:10.1038/nsmb942
Pfund C, Lopez-Hoyo N, Ziegelhoffer T, Schilke BA, Lopez-Buesa P, Walter WA, Wiedmann M, Craig EA (1998) The molecular chaperone Ssb from Saccharomyces cerevisiae is a component of the ribosome-nascent chain complex. EMBO J 17(14):3981–3989. doi:10.1093/emboj/17.14.3981
Conz C, Otto H, Peisker K, Gautschi M, Wolfle T, Mayer MP, Rospert S (2007) Functional characterization of the atypical Hsp70 subunit of yeast ribosome-associated complex. J Biol Chem 282(47):33977–33984. doi:10.1074/jbc.M706737200
Leidig C, Bange G, Kopp J, Amlacher S, Aravind A, Wickles S, Witte G, Hurt E, Beckmann R, Sinning I (2013) Structural characterization of a eukaryotic chaperone-the ribosome-associated complex. Nat Struct Mol Biol 20(1):23–28. doi:10.1038/nsmb.2447
Peisker K, Braun D, Wolfle T, Hentschel J, Funfschilling U, Fischer G, Sickmann A, Rospert S (2008) Ribosome-associated complex binds to ribosomes in close proximity of Rpl31 at the exit of the polypeptide tunnel in yeast. Mol Biol Cell 19(12):5279–5288. doi:10.1091/mbc.E08-06-0661
Yan W, Schilke B, Pfund C, Walter W, Kim S, Craig EA (1998) Zuo, a ribosome-associated DnaJ molecular chaperone. EMBO J 17(16):4809–4817. doi:10.1093/emboj/17.16.4809
Fiaux J, Horst J, Scior A, Preissler S, Koplin A, Bukau B, Deuerling E (2010) Structural analysis of the ribosome-associated complex (RAC) reveals an unusual Hsp70/Hsp40 interaction. J Biol Chem 285(5):3227–3234. doi:10.1074/jbc.M109.075804
Kim SY, Craig EA (2005) Broad sensitivity of Saccharomyces cerevisiae lacking ribosome-associated chaperone ssb or zuo1 to cations, including aminoglycosides. Eukaryot Cell 4(1):82–89. doi:10.1128/EC.4.1.82-89.2005
Nelson RJ, Ziegelhoffer T, Nicolet C, Werner-Washburne M, Craig EA (1992) The translation machinery and 70 kd heat shock protein cooperate in protein synthesis. Cell 71(1):97–105
Hundley H, Eisenman H, Walter W, Evans T, Hotokezaka Y, Wiedmann M, Craig E (2002) The in vivo function of the ribosome-associated Hsp70, Ssz1, does not require its putative peptide-binding domain. Proc Natl Acad Sci U S A 99(7):4203–4208. doi:10.1073/pnas.062048399
Rauch T, Hundley HA, Pfund C, Wegrzyn RD, Walter W, Kramer G, Kim SY, Craig EA, Deuerling E (2005) Dissecting functional similarities of ribosome-associated chaperones from Saccharomyces cerevisiae and Escherichia coli. Mol Microbiol 57(2):357–365. doi:10.1111/j.1365-2958.2005.04690.x
Willmund F, del Alamo M, Pechmann S, Chen T, Albanese V, Dammer EB, Peng J, Frydman J (2013) The cotranslational function of ribosome-associated Hsp70 in eukaryotic protein homeostasis. Cell 152(1-2):196–209. doi:10.1016/j.cell.2012.12.001
Shulga N, James P, Craig EA, Goldfarb DS (1999) A nuclear export signal prevents Saccharomyces cerevisiae Hsp70 Ssb1p from stimulating nuclear localization signal-directed nuclear transport. J Biol Chem 274(23):16501–16507
Zhang S, Lockshin C, Herbert A, Winter E, Rich A (1992) Zuo, a putative Z-DNA binding protein in Saccharomyces cerevisiae. EMBO J 11(10):3787–3796
Hundley HA, Walter W, Bairstow S, Craig EA (2005) Human Mpp11 J protein: ribosome-tethered molecular chaperones are ubiquitous. Science 308(5724):1032–1034. doi:10.1126/science.1109247
Jaiswal H, Conz C, Otto H, Wolfle T, Fitzke E, Mayer MP, Rospert S (2011) The chaperone network connected to human ribosome-associated complex. Mol Cell Biol 31(6):1160–1173. doi:10.1128/MCB.00986-10
Otto H, Conz C, Maier P, Wolfle T, Suzuki CK, Jeno P, Rucknagel P, Stahl J, Rospert S (2005) The chaperones MPP11 and Hsp70L1 form the mammalian ribosome-associated complex. Proc Natl Acad Sci U S A 102(29):10064–10069. doi:10.1073/pnas.0504400102
Albanese V, Reissmann S, Frydman J (2010) A ribosome-anchored chaperone network that facilitates eukaryotic ribosome biogenesis. J Cell Biol 189(1):69–81. doi:10.1083/jcb.201001054
Henras AK, Soudet J, Gerus M, Lebaron S, Caizergues-Ferrer M, Mougin A, Henry Y (2008) The post-transcriptional steps of eukaryotic ribosome biogenesis. Cell Mol Life Sci 65(15):2334–2359. doi:10.1007/s00018-008-8027-0
von Plehwe U, Berndt U, Conz C, Chiabudini M, Fitzke E, Sickmann A, Petersen A, Pfeifer D, Rospert S (2009) The Hsp70 homolog Ssb is essential for glucose sensing via the SNF1 kinase network. Genes Dev 23(17):2102–2115. doi:10.1101/gad.529409
Ducett JK, Peterson FC, Hoover LA, Prunuske AJ, Volkman BF, Craig EA (2013) Unfolding of the C-terminal domain of the J-protein Zuo1 releases autoinhibition and activates Pdr1-dependent transcription. J Mol Biol 425(1):19–31. doi:10.1016/j.jmb.2012.09.020
Prunuske AJ, Waltner JK, Kuhn P, Gu B, Craig EA (2012) Role for the molecular chaperones Zuo1 and Ssz1 in quorum sensing via activation of the transcription factor Pdr1. Proc Natl Acad Sci U S A 109(2):472–477. doi:10.1073/pnas.1119184109
Eisenman HC, Craig EA (2004) Activation of pleiotropic drug resistance by the J-protein and Hsp70-related proteins, Zuo1 and Ssz1. Mol Microbiol 53(1):335–344. doi:10.1111/j.1365-2958.2004.04134.x
Olzscha H, Schermann SM, Woerner AC, Pinkert S, Hecht MH, Tartaglia GG, Vendruscolo M, Hayer-Hartl M, Hartl FU, Vabulas RM (2011) Amyloid-like aggregates sequester numerous metastable proteins with essential cellular functions. Cell 144(1):67–78. doi:10.1016/j.cell.2010.11.050
Tyedmers J, Mogk A, Bukau B (2010) Cellular strategies for controlling protein aggregation. Nat Rev Mol Cell Biol 11(11):777–788. doi:10.1038/nrm2993
Kirstein-Miles J, Morimoto RI (2013) Ribosome-associated chaperones act as proteostasis sentinels. Cell Cycle 12(15):2335–2336. doi:10.4161/cc.25703
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Scior, A., Deuerling, E. (2014). Functions of Ribosome-Associated Chaperones and their Interaction Network. In: Houry, W. (eds) The Molecular Chaperones Interaction Networks in Protein Folding and Degradation. Interactomics and Systems Biology, vol 1. Springer, New York, NY. https://doi.org/10.1007/978-1-4939-1130-1_2
Download citation
DOI: https://doi.org/10.1007/978-1-4939-1130-1_2
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4939-1129-5
Online ISBN: 978-1-4939-1130-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)