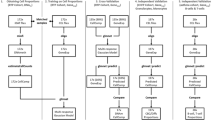
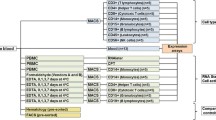
Abstract
Since most immunological and hematological conditions might be expected to alter whole blood gene expression, its examination can lead to insights into disease processes, and biomarkers to assess molecular phenotypes, disease states, progression and response to therapy. In this chapter we describe collection and storage of RNA from whole blood, techniques to measure gene expression, and analytical approaches to identify the dysregulated gene expression using pathway and clustering analysis, gene set enrichment, heat map approaches, and cell subset deconvolution.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Pham MX, Teuteberg JJ, Kfoury AG et al (2010) Gene-expression profiling for rejection surveillance after cardiac transplantation. N Engl J Med 362:1890–1900
Lansky A, Elashoff MR, Ng V et al (2012) A gender-specific blood-based gene expression score for assessing obstructive coronary artery disease in nondiabetic patients: results of the Personalized Risk Evaluation and Diagnosis in the Coronary Tree (PREDICT) trial. Am Heart J 164:320–326
Kirou KA, Gkrouzman E (2013) Anti-interferon alpha treatment in SLE. Clin Immunol 148:303–312
McKay F, Schibeci S, Heard R et al (2006) Analysis of neutralizing antibodies to therapeutic interferon-beta in multiple sclerosis patients: a comparison of three methods in a large Australasian cohort. J Immunol Methods 310: 20–29
Parnell GP, McLean AS, Booth DR et al (2012) A distinct influenza infection signature in the blood transcriptome of patients with severe community-acquired pneumonia. Crit Care 16:R157
Parnell G, McLean A, Booth D et al (2011) Aberrant cell cycle and apoptotic changes characterise severe influenza A infection–a meta-analysis of genomic signatures in circulating leukocytes. PLoS One 6:e17186
Le-Niculescu H, Levey DF, Ayalew M et al (2013) Discovery and validation of blood biomarkers for suicidality. Mol Psychiatry 18:1249–1264
Parnell GP, Tang BM, Nalos M et al (2013) Identifying key regulatory genes in the whole blood of septic patients to monitor underlying immune dysfunctions. Shock 40:166–174
Debey-Pascher S, Hofmann A, Kreusch F et al (2011) RNA-stabilized whole blood samples but not peripheral blood mononuclear cells can be stored for prolonged time periods prior to transcriptome analysis. J Mol Diagn 13:452–460
Joehanes R, Johnson AD, Barb JJ et al (2012) Gene expression analysis of whole blood, peripheral blood mononuclear cells, and lymphoblastoid cell lines from the Framingham Heart Study. Physiol Genomics 44:59–75
Whitney AR, Diehn M, Popper SJ et al (2003) Individuality and variation in gene expression patterns in human blood. Proc Natl Acad Sci U S A 100:1896–1901
Maes M, Stevens W, Scharpe S et al (1994) Seasonal variation in peripheral blood leukocyte subsets and in serum interleukin-6, and soluble interleukin-2 and -6 receptor concentrations in normal volunteers. Experientia 50: 821–829
Debey S, Schoenbeck U, Hellmich M et al (2004) Comparison of different isolation techniques prior gene expression profiling of blood derived cells: impact on physiological responses, on overall expression and the role of different cell types. Pharmacogenomics J 4:193–207
Reimann KA, Chernoff M, Wilkening CL et al (2000) Preservation of lymphocyte immunophenotype and proliferative responses in cryopreserved peripheral blood mononuclear cells from human immunodeficiency virus type 1-infected donors: implications for multicenter clinical trials. The ACTG Immunology Advanced Technology Laboratories. Clin Diagn Lab Immunol 7:352–359
Costantini A, Mancini S, Giuliodoro S et al (2003) Effects of cryopreservation on lymphocyte immunophenotype and function. J Immunol Methods 278:145–155
Mollinedo F, Lopez-Perez R, Gajate C (2008) Differential gene expression patterns coupled to commitment and acquisition of phenotypic hallmarks during neutrophil differentiation of human leukaemia HL-60 cells. Gene 419: 16–26
Ozsolak F, Milos PM (2011) RNA sequencing: advances, challenges and opportunities. Nat Rev Genet 12:87–98
Kruhoffer M, Dyrskjot L, Voss T et al (2007) Isolation of microarray-grade total RNA, microRNA, and DNA from a single PAXgene blood RNA tube. J Mol Diagn 9:452–458
Lander ES, Waterman MS (1988) Genomic mapping by fingerprinting random clones: a mathematical analysis. Genomics 2:231–239
Trapnell C, Pachter L, Salzberg SL (2009) TopHat: discovering splice junctions with RNA-Seq. Bioinformatics 25:1105–1111
Deng N, Puetter A, Zhang K et al (2011) Isoform-level microRNA-155 target prediction using RNA-seq. Nucleic Acids Res 39:e61
Xu G, Deng N, Zhao Z et al (2011) SAMMate: a GUI tool for processing short read alignments in SAM/BAM format. Source Code Biol Med 6:2
Bolstad BM, Irizarry RA, Astrand M et al (2003) A comparison of normalization methods for high density oligonucleotide array data based on variance and bias. Bioinformatics 19:185–193
Robinson MD, McCarthy DJ, Smyth GK (2010) edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics 26:139–140
Anders S, Huber W (2010) Differential expression analysis for sequence count data. Genome Biol 11:R106
Trapnell C, Williams BA, Pertea G et al (2010) Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 28:511–515
Trapnell C, Roberts A, Goff L et al (2012) Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat Protoc 7:562–578
Oshlack A, Robinson MD, Young MD (2010) From RNA-seq reads to differential expression results. Genome Biol 11:220
Slonim DK (2002) From patterns to pathways: gene expression data analysis comes of age. Nat Genet 32(Suppl):502–508
Ogata H, Goto S, Sato K et al (1999) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 27:29–34
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet 25:25–29
Subramanian A, Tamayo P, Mootha VK et al (2005) Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A 102:15545–15550
Shannon W, Culverhouse R, Duncan J (2003) Analyzing microarray data using cluster analysis. Pharmacogenomics 4:41–52
Eisen MB, Spellman PT, Brown PO et al (1998) Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci U S A 95:14863–14868
Hartigan JA, Wong MA (1979) Algorithm AS 136: A K-means clustering algorithm. J Roy Stat Soc C Appl Stat 28:100–108
Kohonen T (1990) The self-organizing map. Proc IEEE 78:1464–1480
Abbas AR, Wolslegel K, Seshasayee D et al (2009) Deconvolution of blood microarray data identifies cellular activation patterns in systemic lupus erythematosus. PLoS One 4:e6098
Zhong Y, Liu Z (2012) Gene expression deconvolution in linear space. Nat Methods 9:8–9, author reply 9
Abbas AR, Baldwin D, Ma Y et al (2005) Immune response in silico (IRIS): immune-specific genes identified from a compendium of microarray expression data. Genes Immun 6:319–331
Heng TS, Painter MW (2008) The immunological genome project: networks of gene expression in immune cells. Nat Immunol 9:1091–1094
Gandhi KS, McKay FC, Cox M et al (2010) The multiple sclerosis whole blood mRNA transcriptome and genetic associations indicate dysregulation of specific T cell pathways in pathogenesis. Hum Mol Genet 19: 2134–2143
Cox MB, Cairns MJ, Gandhi KS et al (2010) MicroRNAs miR-17 and miR-20a inhibit T cell activation genes and are under-expressed in MS whole blood. PLoS One 5:e12132
Quail MA, Smith M, Coupland P et al (2012) A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics 13:341
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this protocol
Cite this protocol
Parnell, G.P., Booth, D.R. (2014). Whole Blood Transcriptomic Analysis to Identify Clinical Biomarkers of Drug Response. In: Yan, Q. (eds) Pharmacogenomics in Drug Discovery and Development. Methods in Molecular Biology, vol 1175. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-0956-8_3
Download citation
DOI: https://doi.org/10.1007/978-1-4939-0956-8_3
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-0955-1
Online ISBN: 978-1-4939-0956-8
eBook Packages: Springer Protocols