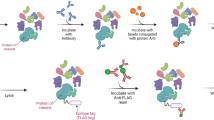
Abstract
Proteins play important roles in biochemical processes. Most biological functions are realized through protein–protein interactions (PPI). Co-immunoprecipitation is the most straightforward method to detect PPI. With the development of modern mass spectrometry (MS), throughput, sensitivity, and confidence for the detection of PPI can be readily achieved by scaling up traditional antibody-based strategies. Herein, we describe a typical workflow for general PPI detection using mass spectrometry coupled techniques, covering from Co-immunoprecipitation (Co-IP), to gel display, in-gel digestion, liquid chromatography mass spectrometry (LC-MS) analysis, as well as result interpretation and statistic filtering. This protocol provides an overview of the technique as well as practical tips.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Young KH (1998) Yeast two-hybrid: so many interactions, (in) so little time. Biol Reprod 58(2):302–311
Bonetta L (2005) The inside scoop[mdash]evaluating gene delivery methods. Nat Meth 2(11):875–883
Gavin AC et al (2002) Functional organization of the yeast proteome by systematic analysis of protein complexes. Nature 415(6868):141–147
John RY III (1998) Database searching using mass spectrometry data. Electrophoresis 19(6):893–900
Craig R, Beavis RC (2004) TANDEM: matching proteins with tandem mass spectra. Bioinformatics 20(9):1466–1467
Perkins DN, Pappin DJ, Creasy DM, Cottrell JS (1999) Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20(18):3551–3567
Liu G, et al. (2012) Using ProHits to store, annotate, and analyze affinity purification-mass spectrometry (AP-MS) data. Curr Protoc Bioinformatics Chapter 8:16
Liu G et al (2010) ProHits: integrated software for mass spectrometry-based interaction proteomics. Nat Biotechnol 28(10):1015–1017
Choi H et al (2012) SAINT-MS1: protein–protein interaction scoring using label-free intensity data in affinity purification-mass spectrometry experiments. J Proteome Res 11(4):2619–2624
Choi H et al (2011) SAINT: probabilistic scoring of affinity purification-mass spectrometry data. Nat Methods 8(1):70–73
Choi H, et al. (2012) Analyzing protein–protein interactions from affinity purification-mass spectrometry data with SAINT. Curr Protoc Bioinformatics Chapter 8:15
Shevchenko A et al (2006) In-gel digestion for mass spectrometric characterization of proteins and proteomes. Nat Protoc 1(6):2856–2860
Elias JE, Gygi SP (2010) Target-decoy search strategy for mass spectrometry-based proteomics. Methods Mol Biol 604:55–71
Lavallee-Adam M et al (2011) Modeling contaminants in AP-MS/MS experiments. J Proteome Res 10(2):886–895
Acknowledgements
D.F. holds a Canada Research Chair Tier I in Proteomics and Systems Biology. This work was supported by NSERC and CIHR.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this protocol
Cite this protocol
Ning, Z., Hawley, B., Chiang, CK., Seebun, D., Figeys, D. (2014). Detecting Protein–Protein Interactions/Complex Components Using Mass Spectrometry Coupled Techniques. In: Miyamoto-Sato, E., Ohashi, H., Sasaki, H., Nishikawa, Ji., Yanagawa, H. (eds) Transcription Factor Regulatory Networks. Methods in Molecular Biology, vol 1164. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4939-0805-9_1
Download citation
DOI: https://doi.org/10.1007/978-1-4939-0805-9_1
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4939-0804-2
Online ISBN: 978-1-4939-0805-9
eBook Packages: Springer Protocols