Abstract
The existence of citrulline in proteins was first described in the skin. In a paper published in Nature in 1958, George Rogers reported citrulline in a protein of hair follicles (Rogers and Simmonds 1958). Twenty-eight years later, Rothnagel and Rogers purified and characterised the corresponding protein and called it trichohyalin (Rothnagel and Rogers 1986). Since then, deiminated proteins have been detected in almost all cells, tissues and organs. The enzymes responsible for this posttranslational modification, the peptidylarginine deiminases (PADs), also known as protein-arginine deiminases, are becoming increasingly well known. Five types of PADs have been identified in humans and other mammals, the PAD1, 2, 3, 4 (also known as PAD5), and 6 (Vossenaar et al. 2003; Chavanas et al. 2004; Balandraud et al. 2005). They are encoded by five paralogous genes clustered on chromosome 1p35-36 and named PADI1, 2, 3, 4 and 6 (Vossenaar et al. 2003; Chavanas et al. 2004). The importance of PADs in many cellular processes is now recognised (Klose and Zhang 2007; Li et al. 2010; Struyf et al. 2009; Esposito et al. 2007) and PADs have been involved in the pathogenesis of autoimmune diseases, e.g. rheumatoid arthritis (Klareskog et al. 2008; Sebbag et al. 2004), multiple sclerosis (Harauz and Musse 2007; Kim et al. 2003) and cancer (Slack et al. 2011a), that are described in more detail in other chapters of this book. Here, we report on the location of PADs expressed in skin; the mechanisms involved in the regulation of their expression and activity in keratinocytes, their skin targets and physiological roles; and, finally, their possible contribution to skin diseases.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Peptidylarginine deiminase
- Keratinocyte
- Immunohistochemistry
- Localization
- Gene regulation
- Differentiation
- Skin
7.1 Introduction
The existence of citrulline in proteins was first described in the skin. In a paper published in Nature in 1958, George Rogers reported citrulline in a protein of hair follicles (Rogers and Simmonds 1958). Twenty-eight years later, Rothnagel and Rogers purified and characterised the corresponding protein and called it trichohyalin (Rothnagel and Rogers 1986). Since then, deiminated proteins have been detected in almost all cells, tissues and organs. The enzymes responsible for this posttranslational modification, the peptidylarginine deiminases (PADs), also known as protein-arginine deiminases, are becoming increasingly well known. Five types of PADs have been identified in humans and other mammals, the PAD1, 2, 3, 4 (also known as PAD5), and 6 (Vossenaar et al. 2003; Chavanas et al. 2004; Balandraud et al. 2005). They are encoded by five paralogous genes clustered on chromosome 1p35-36 and named PADI1, 2, 3, 4 and 6 (Vossenaar et al. 2003; Chavanas et al. 2004). The importance of PADs in many cellular processes is now recognised (Klose and Zhang 2007; Li et al. 2010; Struyf et al. 2009; Esposito et al. 2007) and PADs have been involved in the pathogenesis of autoimmune diseases, e.g. rheumatoid arthritis (Klareskog et al. 2008; Sebbag et al. 2004), multiple sclerosis (Harauz and Musse 2007; Kim et al. 2003) and cancer (Slack et al. 2011a), that are described in more detail in other chapters of this book. Here, we report on the location of PADs expressed in skin; the mechanisms involved in the regulation of their expression and activity in keratinocytes, their skin targets and physiological roles; and, finally, their possible contribution to skin diseases.
The skin provides mechanical protection to the organism and is an important barrier for preventing the invasion of pathogens, the entry of exogenous substances including allergens and toxins, and the uncontrolled loss of body water and solutes. This so-called barrier function is performed by the epidermis (Madison 2003), a stratified squamous epithelium mainly composed of keratinocytes.
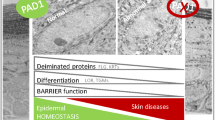
Terminal differentiation of keratinocytes is an oriented and complex program of gene expression from the proliferative basal layer of the epidermis to the upper horny layer, also named the stratum corneum (Fig. 7.1a). During their journey through the spinous and granular layers, the cells sequentially turn specific genes on and off and undergo a series of structural and metabolic modifications. For instance, the expression of keratins KRT1 and KRT10 starts in the spinous layer, whereas filaggrin (FLG) is detected from the granular layer. Finally, the granular keratinocytes undergo a specialised form of programmed cell death called cornification. Cornification is characterised by (1) the elimination of all organelles and the nucleus; (2) the formation of a resistant and insoluble protein shell at the keratinocyte periphery, the cornified cell envelope; (3) the transformation of desmosomes, the intercellular junctional structures, into corneodesmosomes; and (4) the aggregation of the keratin intermediate filaments to form a macrofibrillar intracellular matrix. The resulting corneocytes are embedded in lipidic lamellae and form the thin, highly organised and resilient horny layer (Madison 2003; Candi et al. 2005). In order to maintain the thickness of the horny layer, the upper corneocytes detach from the skin surface during the strongly controlled process of desquamation and are replaced by newly differentiated cells. When human and rodent skin was probed with the anti-modified citrulline antibodies developed by Tatsuo Senshu (Yokohama, Japan), only the horny layer was stained (Senshu et al. 1996) (Fig. 7.1b).
Deiminated human epidermal proteins: involvement of PADs in the hydration of the stratum corneum. (a) The four epidermal keratinocyte layers are shown on a stained section of human skin (center): the stratum corneum (SC), granulosum (SG), spinosum (SP) and basale (SB). The immunodetection patterns of keratin (K) 1 and K10, profilaggrin and filaggrin ((pro)FLG), deiminated keratins (dK) and filaggrin (dFLG), and PADs in human epidermis are schematically indicated on the left. The complex metabolism of profilaggrin is schematically represented on the right: deimination of filaggrin, performed by PAD1 and PAD3, is essential for the degradation of the protein to free amino acids and the production of the natural moisturizing factor. (b) Immunochemical detection of deiminated proteins (in brown) in the stratum corneum of human epidermis. Scale bar = 40 μm
7.2 PADs Expressed in Skin
7.2.1 In the Epidermis
The expression of only three PADI genes, i.e. PADI1, 2 and 3 (Guerrin et al. 2003; Nachat et al. 2005a), has been evidenced in human skin and epidermis by RT-PCR experiments. In cultured human primary keratinocytes, mRNAs encoding the same three PAD isotypes, but not PAD4 or PAD6, have also been detected (Méchin et al. 2010). In agreement with these findings, only PAD1, 2 and 3 have been immunodetected on skin sections with anti-peptide antibodies specific for each of the isoforms. PAD1 has been localised in the cytoplasm of keratinocytes throughout the whole human epidermis, with a higher expression in the granular cells and in the corneocyte intracellular matrix. In the granular cells, it is associated with keratohyalin granules and with keratin intermediate filaments. PAD2 has been detected in the cytoplasm of spinous keratinocytes and, with a more intense staining, at the periphery of granular keratinocytes. PAD3 is located in keratohyalin granules, in the cytoplasm of granular keratinocytes and in the matrix of the lower corneocytes (Figs. 7.1a and 7.2a). Immunoblotting experiments have confirmed these data. In particular, immunoblotting carried out on samples obtained from the superficial horny layer using adhesive tape stripping has evidenced that PAD1 is the only PAD isotype present in the upper corneocytes. The same pattern of PAD expression has been described in mouse epidermis (Guerrin et al. 2003; Nachat et al. 2005a; Méchin et al. 2005; Coudane et al. 2011). In addition, the expression of PAD4 in rat epidermis has also been reported (Ishigami et al. 2001).
Immunodetection of PAD3 in the epidermis and inner root sheath of hair follicles. Cryosections of human skin were analysed by confocal microscopy with anti-PAD3 rabbit antibodies, with a monoclonal antibody directed against profilaggrin and filaggrin ((pro) filaggrin) and with a monoclonal antibody specific for corneodesmosin. (a) In the epidermis, PAD3 is co-located with profilaggrin in the cytoplasm of the upper granular keratinocytes and with filaggrin in the lower corneocytes. (b) In the hair follicles, PAD3 is detected in the cytoplasm of keratinocytes in the Huxley layer of the inner root sheath. Bar: 10 μm
7.2.2 In Skin Appendages
PAD1 and PAD3 have been immunodetected in the concentric epithelial sheaths forming hair follicles at the anagen stage (Nachat et al. 2005b). PAD1 is expressed by differentiated keratinocytes, first those of the cuticle and Huxley’s layer of the inner root sheath and second those of the companion layer between the inner and the outer root sheaths. PAD3 is expressed in cells of the inner root sheath (Fig. 7.2b) and the medulla. Both enzymes have been immunodetected in the keratinocyte cytoplasm.
PAD1 and PAD2 have also been observed in the secretory and myoepithelial cells of the sweat glands and in the arrector pili muscles (Nachat et al. 2005b). So far, no PADs have been detected in human sebaceous glands.
7.3 Regulation of PAD Expression in Keratinocytes
7.3.1 Minimal Promoters of PADI Genes and Bound Transcription Factors
Most of the genes expressed during the program of keratinocyte differentiation are regulated at the transcriptional level. This is probably also the case for the PADI genes. When differentiation of cultured human epidermal keratinocytes is induced by treatment with 10−7 M of 1-α,25-dihydroxy-vitamin D3 (VitD) for 24 h, higher amounts of mRNAs encoding PAD1 (~3-fold increase), PAD2 (~8-fold) and PAD3 (~10-fold) are detected (Méchin et al. 2010). Expression of PADI1, 2 and 3 mRNAs is also improved when the extracellular calcium concentration is increased (Méchin et al. 2010; Chavanas et al. 2008; Dong et al. 2005), another well-known way of inducing the differentiation of keratinocytes.
The minimal promoters of the PADI1, PADI2 and PADI3 genes have recently been delineated as short sequences of 195, 132 and 129 base pairs, respectively, upstream of the transcription initiation site (Dong et al. 2005, 2006, 2008). Electrophoretic mobility-shift assays, chromatin immunoprecipitation and small interfering RNA experiments have shown that binding of transcription factors of the ubiquitous stimulator protein (Sp) family, namely, Sp1 and Sp3, is crucial for the activity of these proximal promoters in keratinocytes (Dong et al. 2005, 2006, 2008). This is not surprising, since functional binding sites for Sp1 are frequently found in the promoter regions of genes expressed during the late steps of keratinocyte differentiation. These include the genes encoding keratin 1, transglutaminases 1 and 3, cornified cell envelope components and profilaggrin (Lee et al. 1996; Wong et al. 2005; Jang and Steinert 2002; Crish et al. 2006; Eckert et al. 2004; Markova et al. 2007).
The additional binding of MZF1 and NF-Y transcription factors is necessary to regulate the expression of PADI1 and PADI3, respectively (Dong et al. 2006, 2008). Also, the expression of both Sp1 and MZF1 is increased after calcium stimulation of keratinocyte differentiation (Dong et al. 2008; Wong et al. 2005). Interestingly, Sp1 and MZF1 are also involved in the regulation of the BLMH gene (Kamata et al. 2011). This gene encodes a neutral cysteine protease, bleomycin hydrolase, implicated downstream of PAD1 and PAD3 in the processing of FLG (see Sect. 5.2). This suggests a possible co-regulation of FLG processing-related enzymes. In addition, a proximal TATA-box is present in PAD1 and PAD3, but not PAD2, genes (Dong et al. 2005, 2006, 2008).
Since binding sites for Sp1, MZF1 and NF-Y have been identified in silico upstream of the transcription start site of the mouse orthologous genes (Padi1, Padi2 and Padi3) (Dong et al. 2005, 2006, 2008), the same transcription factors are probably involved in the regulation of both human and mouse PAD genes. As in humans, when wild-type, but not VitD receptor null, mouse keratinocytes are treated with an analogue of VitD (EB1089), Padi3 is up-regulated. This twofold activation occurs through the binding of activated VitD receptor–β-catenin complexes to two VitD response elements located ~3 kb upstream of the transcription initiation site (Pálmer et al. 2008).
7.3.2 Role of Non-coding Conserved Sequences
However, considered alone, Sp1 and NF-Y binding to its proximal promoter cannot explain the tight control of PADI3 expression in the granular keratinocytes, since these transcription factors are also involved in the regulation of PADI4 (Dong et al. 2007), the expression of which has not been detected in the keratinocytes. Therefore, other levels of PADI3 control have been suspected, in particular the role of non-coding evolutionarily conserved sequences.
The first to be identified was an 8-kb region located between PADI2 and PADI1, 42 kb upstream of PADI2 and 37 kb upstream of PADI1 (the two genes are in the opposite transcription orientation). This sequence groups together many potential transcription factor-binding sites and is in an open conformation state of the chromatin in differentiated keratinocytes. It is therefore likely to bind transcriptional activators (Chavanas et al. 2004, 2008). In addition, by the chromosome conformation capture technique, it has been shown to physically interact with the PADI3 promoter in the nuclei of differentiating keratinocytes through a chromatin loop spanning 86 kb (Chavanas et al. 2008). Several long-range enhancers and bound activators have been experimentally recognised in this region. Two segments of 346 and 245 bp, 1 kb distant from each other, cooperate in calcium-differentiated epidermal keratinocytes to enhance the activity of the PADI3 gene minimal promoter located 82 kb away (Adoue et al. 2008). They have no effect when tested independently but act in an orientation-independent and copy number-dependent manner. Their effect has not been observed in proliferative epidermal keratinocytes, in human fibroblasts or cervix adenocarcinoma HeLa cells or on the PADI2 minimal promoter. This strongly suggests that these two segments, called PAD intergenic enhancer segment 1 (PIE-S1) and PIE-S2, form a real PADI3 bipartite enhancer. PIE-S2 binds distinct transcription factors of the AP-1 family according to the differentiation state of keratinocytes, junD homodimer in proliferative cells and c-Jun homodimer in differentiated cells (Adoue et al. 2008). PIE-S1 contains an MIBP1/RFX1-binding site (Adoue et al. 2008), but binding of either of these two transcription factors has not yet been proved.
An additional non-coding conserved segment of 63 bp, called PIE, has also been shown to display strong enhancer activity on the PADI3 gene minimal promoter in calcium-differentiated keratinocytes (Chavanas et al. 2008). The enhancer activity of PIE does not depend on its orientation; it is low in proliferative keratinocytes and insignificant in HaCaT and Hela cells. It is low on the PADI2 minimal promoter and null on PADI1 and PADI4 promoter. To be active, PIE requires the binding of c-Jun and c-Fos, another transcription factor of the AP-1 family. In addition, PIE seems to interact functionally with the two CAAT boxes of the PADI promoter, probably through a direct interaction between c-Jun and NF-Y (Chavanas et al. 2008).
Long-range regulatory elements are important for the coordinated regulation of many clustered genes, at distances of up to 1 Mb from their cognate promoters and in several cell types (Li et al. 2002). Whether this is also a key mechanism in regulating genes essential for terminally differentiated keratinocytes is less certain. However, a network of conserved non-coding sequences involved in the regulation of the numerous genes of the so-called epidermal differentiation complex in keratinocytes has been described recently (Martin et al. 2004; de Guzman Strong et al. 2010). The expression of the p63 gene is also controlled by a long-range keratinocyte-specific enhancer (Antonini et al. 2006).
When orthologous PADI genes from multiple mammalian species are aligned, a highly conserved 1 kb region is revealed in the PADI1 first intron, suggestive of a biological role (Ying et al. 2010). A 267 bp fragment of this region has been shown to enhance the activity of the PADI1 minimal promoter in an orientation-independent manner in both proliferative and calcium-differentiated human keratinocytes. Binding of p65 and p50 subunits of NF-kappaB transcription factor is necessary for this enhancer activity. A physical interaction between the PADI1 minimal promoter and these intronic conserved non-coding sequences, located 2.2 kb apart, has been evidenced, indicating chromatin looping (Ying et al. 2010). NF-kappaB involvement in keratinocyte differentiation and senescence is well known, as is its transcriptional effect via direct binding to response elements located in gene introns, including c-Fos (Bernard et al. 2004; Bell et al. 2003; Charital et al. 2009).
As a whole, these data show that the transcription of PADI gene during keratinocyte differentiation is under the control of multiple and complex regulatory mechanisms, including chromatin structure remodelling (Fig. 7.3).
Schematic model of the transcriptional regulation of PADI1 and PADI3 gene expression in human keratinocytes. Part of the PADI gene locus is located in shown: the 5′ region of PADI2, the intergenic region including two long-distance enhancers (namely, PIE and PIES1/S2), the entire PADI1 gene including the conserved non-coding sequence its first intron (CNSi) and the 5′ region of PADI3. Minimal promoters are indicated by coloured rectangular boxes and the orientation of transcription by black arrows. Bound transcription factors are shown, as are their binding sites. To allow activation of the PADI3 promoter by PIE and PIES1/S2 enhancers located 87 kb upstream, chromatin has to form a large loop to bring them into physical contact. Similarly, a chromatin loop allows the activation of PADI1 transcription through an interaction between the transcription complex and p50/p65 NF-κB transcription factors bound 2 kb downstream on the CNSi. Transcription factors (MZF1, c-Jun, c-Fos, JunD, NFYA and Sp1/3), the TATA-box-binding protein (TBP) and RNA polymerase II complex (Pol II) are shown as open circles and the transcription factor-binding sites by open boxes. Note that the distances (in bases) are not drawn to scale
7.3.3 Regulation at the Translational Level
As previously reported for other tissues or cells, including optic nerve cells (Bhattacharya et al. 2006) and monocytes (Vossenaar et al. 2004), PAD expression in keratinocytes seems to be also regulated at the translational level. For example, treatment of keratinocytes with VitD strongly increases the amount of PAD1, PAD2 and PAD3 mRNA but has no effect on the corresponding protein (Méchin et al. 2010).
7.4 Regulation of PAD Activity in Keratinocytes
The presence of one PAD in keratinocytes at a particular time does not necessarily mean that the deimination of proteins takes place at the same time. Although PADs are immunodetected in basal and suprabasal living epidermal keratinocytes, deiminated proteins are only immunostained in corneocytes. In addition, even though Hela and HaCaT cells express at least one PAD, we have not been able to detect deiminated proteins, even in cells cultured at a high extracellular calcium concentrations (Méchin et al. 2010). In the same way, no deiminated proteins have been detected in cells of sweat glands and arrector muscles, where PAD1 and PAD2 are expressed (Nachat et al. 2005b). We suspect that the local intracellular concentration of calcium plays a role in controlling PAD activity, this ion being required for their activity in vitro.
Auto-deimination of PADs may also be involved in the regulation of their activity. We have observed calcium-dependent auto-deimination of PAD1, 2 and 3 during in vitro incubations. This modification reduces, but does not suppress, their activity and changes PAD3 structure. In particular, the distances between the four major amino acids of the active site increase (Méchin et al. 2010). Considering the high sequence homologies between the three isotypes, this is probably true for PAD1 and PAD2 as well. Similarly, PAD4 is also auto-deiminated in vitro and in vivo in activated neutrophils. This posttranslational modification changes the structure of the enzyme and could either inactivate it or modulate its ability to interact with other histone-modifying enzymes (Slack et al. 2011b; Andrade et al. 2010).
7.5 Deiminated Proteins and Role of Deimination in Skin
7.5.1 In the Appendages
As already mentioned in the introduction (Sect. 1) to this chapter, the first protein shown to be deiminated was trichohyalin. Trichohyalin is a member of the S100 fused-type protein (SFTP) family (Henry et al. 2012). Like the other SFTPs, trichohyalin is a large protein (220 kDa in human) formed by three domains: an amino-terminal domain homologous to S100A proteins and containing two functional EF-hand calcium-binding sites (91 amino acids long), a highly charged central domain formed by a series of peptide repeats (1,581 amino acids long) and a short carboxy-terminal tail (50 amino acids long). Trichohyalin is preferentially expressed in the inner root sheath and medulla of hair follicles and in the granular layer of the epithelium of dorsal tongue papillae (Hamilton et al. 1991; O’Keefe et al. 1993). It initially accumulates as large cytoplasmic granules. Then it associates with keratin intermediate filaments. Later it becomes cross-linked to itself, to keratin head and tail domains and to several cornified cell envelope components through ε-(γ-glutamyl) lysine isodipeptide bonds catalysed by transglutaminases (O’Keefe et al. 1993). Trichohyalin serves as a strengthener of the envelopes and as an anchor between the envelope and the corneocyte cytoplasmic matrix. This forms a continuous hardened supramolecular structure conferring high mechanical strength on the inner root sheath (Steinert et al. 2003). Trichohyalin contains 435 arginine residues, many of them being citrullinated. Trichohyalin deimination modifies its α-helical structure, resulting in unfolding; makes it more soluble, inducing granule solubilisation; and makes it a better substrate for transglutamine 3 (Tarcsa et al. 1996, 1997). Because PAD3 and trichohyalin expression patterns are very similar, this isotype is certainly responsible for trichohyalin deimination. In addition, the tail of mouse inner root sheath-specific type-I keratin 27 (formerly K25irs3) and type-II keratin 71 (K6irs1) are also deiminated before the proteins are cross-linked by transglutaminase 3 (Steinert et al. 2003).
S100A3, a calcium- and zinc-binding protein, is another substrate of PAD3 in the hair follicles. S100A3 is located in the cuticle and the cortex of the hair shaft and is believed to be involved in hair shaft formation. In vitro deimination of S100A3 by PAD3 promotes the assembly of a homotetramer and increases its affinity for calcium ions (Kizawa et al. 2008). This is described in greater detail in Chap. 8.
As a whole, these data indicate that deimination in the hair follicles is important for the mechanical resistance of cells in the inner root sheath and hair shaft. No deiminated proteins have yet been identified in sweat gland or arrector muscle cells.
7.5.2 In the Epidermis
Several epidermal deiminated proteins have been characterized, and all of them are modified in the horny layer (Fig. 7.1a, b). The effect of deimination on their properties is starting to be unravelled and will be described below.
The major targets of PADs in the epidermis are two closely related SFTPs, i.e. FGL and FLG-2 (Henry et al. 2012). Their repetitive central domains show 45 % amino-acid sequence similarity and similar amino-acid compositions with particular high levels of serine, glycine, histidine, glutamine and arginine (70.4 and 74.2 %, respectively, of total amino acids). Both are specifically expressed in the granular layer of the epidermis in the form of large insoluble precursors (400 and 248 kDa) which accumulate in the cytoplasmic keratohyalin granules (Henry et al. 2012; Dale et al. 1990; Hsu et al. 2011). At the stratum granulosum/stratum corneum transition, they are proteolytically processed to smaller basic subunits that interact with and are believed to aggregate keratin intermediate filaments. In the lower stratum corneum, they are co-located in the corneocyte filamentous matrix (Henry et al. 2012; Dale et al. 1990; Hsu et al. 2011). In the upper stratum corneum, FLG and probably FLG-2 subunits are totally degraded by several proteases, including caspase-14, calpain-1 and bleomycin hydrolase (Hsu et al. 2011; Hoste et al. 2011; Kamata et al. 2009; Yamazaki et al. 1997). The resulting amino acids form part of the natural moisturizing factor, a mixture of osmotic molecules allowing water retention in the upper stratum corneum (Harding and Scott 1983; Rawlings and Matts 2005). Some of the amino acids are further modified. For example, trans-urocanic acid, involved in photoprotection since it absorbs part of ultraviolet radiation, is derived from histidine in a reaction catalysed by histidase (Barresi et al. 2011). In contrast, pyrolidone carboxylic acid, the most hygroscopic amino acid, derives from glutamine (Rawlings and Matts 2005). With a pKa of 3.9, it also contributes to the acidification of the superficial stratum corneum. This acidic pH is crucial for the antimicrobial activity of the layer, for its waterproof nature through the control of lipase activities and for the regulation of desquamation (Rawlings and Matts 2005; Harding et al. 2000).
FLG and FLG-2 deimination are thought to be necessary for their dissociation from the matrix. They also promote their proteolysis by calpain-1 and are a requisite for their proteolysis by bleomycin hydrolase (Hsu et al. 2011; Kamata et al. 2009). As a consequence, deimination participates in, and presumably controls, the hydration of the upper epidermis and the epidermal barrier functions. On the basis of their enzymatic properties and their diffuse location within the fibrous matrix of the lower corneocytes (Méchin et al. 2005), PAD1 and PAD3 are probably the isotypes responsible for the deimination of FLG and FLG-2.
In the upper cornified layer, the head and tail of keratin K1 and K10 are deiminated (Senshu et al. 1996; Méchin et al. 2005; Ishida-Yamamoto et al. 2002). The enzyme involved is probably PAD1, since it is the only PAD isoform detected in this location (Méchin et al. 2005). The effect of deimination on the properties of these keratins is not known; however, it is concomitant with the observed ultrastructural modifications of the intracorneocyte fibrous matrix. Similarly to inner root sheath keratins, we could suspect that deimination precedes the cross-linking of these proteins to the cornified cell envelopes.
FLG has been known for a long time. It came back under the spotlight when nonsense mutations of its gene were shown to be responsible for ichthyosis vulgaris (OMIM 146700) and to be a high-risk factor for atopic eczema (OMIM #605803) (Smith et al. 2006; Palmer et al. 2006; Irvine and McLean 2006; Sandilands et al. 2007).
7.6 Deimination and Skin Diseases
Despite the accumulating data obtained on PADs by using skin as a model and the importance of deimination in skin physiology, few data are available concerning PAD implication in skin diseases. Lower amounts of citrullinated keratins have been detected in the epidermis of patients with epidermolytic hyperkeratosis (OMIM #113800; also known as bullous congenital ichthyosiform erythroderma) and psoriasis (Ishida-Yamamoto et al. 2000) (in addition to our unpublished data). However, in one study, paclitaxel, a well-known drug used in cancer therapy, but also an in vitro inhibitor of PAD, has been reported to improve severe psoriasis (Ehrlich et al. 2004). No further data has been published about this topic.
PAD could also be involved in skin tumorigenesis. Differential expression of the four genes encoding PAD1 (PADI1), laminin-γ2 (LAMC2), collagen type IV α1 (COL4A1) and collagen type I α1 (COL1A1) has been claimed as a predictive biomarker of squamous cell carcinomas of the oral cavity and oropharynx (Chen et al. 2008). In a genome-wide study concerning 930 Icelanders with cutaneous basal cell carcinoma, which is the most common cancer among Europeans, a single-nucleotide polymorphism in intron 13 of the PAD6 gene has been identified as a strong genetic risk factor (Stacey et al. 2008). This association has been replicated in an additional population from Eastern Europe. The estimated risk of the mutation carriers is 2.68 times that of noncarriers (Stacey et al. 2008). Finally, expression of PAD4, the isotype involved in gene expression regulation through deimination of histones, has been observed in skin carcinomas and extramammary Paget’s disease (OMIM #167300) (Chang et al. 2009; Urano et al. 1990). Since PAD4 inhibition results in cell cycle arrest and apoptosis (Li et al. 2008) and since PAD4 represses the p53 target genes (Yao et al. 2008), this observation could be of relevance for skin tumours. A more detailed discussion on the role of deimination in cancer is discussed in Chap. 17.
7.7 Conclusion
PADs are increasingly considered as crucial molecular actors in cell physiology and human diseases. The data reported here highlight their importance in skin, particularly in the epidermis and hair follicles. However, more work needs to be done to definitively prove their contribution to skin diseases. Detailed analysis of mechanisms involved in controlling PAD expression and activity during keratinocyte differentiation indicates multiple levels of regulation. This indicates that deimination is a crucial post-translational modification of proteins that require tight control.
References
Adoue V, Chavanas S, Coudane F et al (2008) Long range enhancer differentially regulated by c-jun and junD controls peptidylarginine deiminase-3 gene in keratinocytes. J Mol Biol 384:1048–1055
Andrade F, Darrah E, Gucek M et al (2010) Autocitrullination of human peptidyl arginine deiminase type 4 regulates protein citrullination during cell activation. Arthritis Rheum 62:1630–1640
Antonini D, Rossi B, Han R et al (2006) An autoregulatory loop directs the tissue-specific expression of p63 through a long-range evolutionarily conserved enhancer. Mol Cell Biol 26:3308–3318
Balandraud N, Gouret P, Danchin EG et al (2005) A rigorous method for multigenic families’ functional annotation: the peptidyl arginine deiminase (PADs) proteins family example. BMC Genomics 6:153–157
Barresi C, Stremnitzer C, Mlitz V et al (2011) Increased sensitivity of histidinemic mice to UVB radiation suggests a crucial role of endogenous urocanic acid in photoprotection. J Invest Dermatol 131:188–194
Bell S, Degitz K, Quirling M et al (2003) Involvement of NF-kappaB signalling in skin physiology and disease. Cell Signal 15:1–7
Bernard D, Gosselin K, Monte D et al (2004) Involvement of Rel/nuclear factor-kappaB transcription factors in keratinocyte senescence. Cancer Res 64:472–481
Bhattacharya SK, Crabb JS, Bonilha VL et al (2006) Proteomics implicates peptidyl arginine deiminase 2 and optic nerve citrullination in glaucoma pathogenesis. Invest Ophthalmol Vis Sci 47:2508–2514
Candi E, Schmidt R, Melino G (2005) The cornified envelope: a model of cell death in the skin. Nat Rev Mol Cell Biol 6:328–340
Chang X, Han J, Pang L et al (2009) Increased PADI4 expression in blood and tissues of patients with malignant tumors. BMC Cancer 9:40
Charital YM, van Haasteren G, Massiha A et al (2009) A functional NF-kappaB enhancer element in the first intron contributes to the control of c-fos transcription. Gene 430:116–122
Chavanas S, Méchin M-C, Takahara H et al (2004) Comparative analysis of the mouse and human peptidylarginine deiminase gene clusters reveals highly conserved non-coding segments and a new human gene, PADI6. Gene 330:19–27
Chavanas S, Adoue V, Méchin M-C et al (2008) Long-range enhancer associated with chromatin looping allows AP-1 regulation of the peptidylarginine deiminase 3 gene in differentiated keratinocyte. PLoS One 3(10):e3408
Chen C, Méndez E, Houck J et al (2008) Gene expression profiling identifies genes predictive of oral squamous cell carcinoma. Cancer Epidemiol Biomarkers Prev 17:2152–2162
Coudane F, Mechin M-C, Huchenq A (2011) Deimination and expression of peptidylarginine deiminases during cutaneous wound healing in mice. Eur J Dermatol 21:376–384
Crish JF, Gopalakrishnan R, Bone F et al (2006) The distal and proximal regulatory regions of the involucrin gene promoter have distinct functions and are required for in vivo involucrin expression. J Invest Dermatol 126:305–314
Dale B, Resing KA, Haydock PV (1990) Filaggrins. In: Goldman RD, Steinert PM (eds) Molecular and cellular biology of intermediate filaments. Plenum press, New York, pp 393–412
de Guzman Strong C, Conlan S, Deming CB et al (2010) A milieu of regulatory elements in the epidermal differentiation complex syntenic block: implications for atopic dermatitis and psoriasis. Hum Mol Genet 19:1453–1460
Dong S, Kojima T, Shiraiwa M et al (2005) Regulation of the expression of peptidylarginine deiminase type II gene (PADI2) in human keratinocytes involves Sp1 and Sp3 transcription factors. J Invest Dermatol 124:1026–1033
Dong S, Kanno T, Yamaki A et al (2006) NF-Y and Sp1/Sp3 are involved in the transcriptional regulation of the peptidylarginine deiminase type III gene (PADI3) in human keratinocytes. Biochem J 397:449–459
Dong S, Zhang Z, Takahara H (2007) Estrogen-enhanced peptidylarginine deiminase type IV gene (PADI4) expression in MCF-7 cells is mediated by estrogen receptor-alpha-promoted transfactors activator protein-1, nuclear factor-Y, and Sp1. Mol Endocrinol 21:1617–1629
Dong S, Ying S, Kojima T et al (2008) Crucial roles of MZF1 and Sp1 in the transcriptional regulation of the peptidylarginine deiminase type gene (PADI1) in human keratinocytes. J Invest Dermatol 128:549–557
Eckert RL, Crish JF, Efimova T et al (2004) Regulation of involucrin gene expression. J Invest Dermatol 123:13–22
Ehrlich A, Booher S, Becerra Y et al (2004) Micellar paclitaxel improves severe psoriasis in a prospective phase II pilot study. J Am Acad Dermatol 50:533–540
Esposito G, Vitale AM, Leijten FP et al (2007) Peptidylarginine deiminase (PAD) 6 is essential for oocyte cytoskeletal sheet formation and female fertility. Mol Cell Endocrinol 273:25–31
Guerrin M, Ishigami A, Méchin M-C et al (2003) cDNA cloning, gene organization and expression analysis of human peptidylarginine deiminase type I. Biochem J 370:167–174
Hamilton EH, Jr RE, Hamilton RE, O’Keefe EJ (1991) Trichohyalin: presence in the granular layer and stratum corneum of normal human epidermis. J Invest Dermatol 96:666–672
Harauz G, Musse AA (2007) A tale of two citrullines–structural and functional aspects of myelin basic protein deimination in health and disease. Neurochem Res 32:137–158
Harding CR, Scott IR (1983) Histidine-rich proteins (filaggrins): structural and functional heterogeneity during epidermal differentiation. J Mol Biol 170:651–673. CRC Press LCC, Boca Raton, Florida, USA
Harding CR, Bartolone J, Rawlings AV (2000) Effects of natural moisturizing factor and lactic acid isomers on skin function. In: Loden M, Maibach HI (eds) Dry skin and moisturizers: chemistry and function, CRC Press LCC, Boca Raton, Florida, USA pp 229–241
Henry J, Toulza E, Hsu CY et al (2012) Update on the epidermal differentiation complex. Front Biosci 17:1517–1532
Hoste E, Kemperman P, Devos M et al (2011) Caspase-14 is required for filaggrin degradation to natural moisturizing factors in the skin. J Invest Dermatol 131:2233–2241
Hsu CY, Henry J, Raymond AA et al (2011) Deimination of human filaggrin-2 promotes its proteolysis by calpain 1. J Biol Chem 286:23222–23233
Irvine AD, McLean WH (2006) Breaking the (un)sound barrier: filaggrin is a major gene for atopic dermatitis. J Invest Dermatol 126:1200–1222
Ishida-Yamamoto A, Senshu T, Takahashi H et al (2000) Decreased deiminated keratin K1 in psoriatic hyperproliferative epidermis. J Invest Dermatol 114:701–705
Ishida-Yamamoto A, Senshu T, Eady RA et al (2002) Sequential reorganization of cornified cell keratin filaments involving filaggrin-mediated compaction and keratin 1 deimination. J Invest Dermatol 118:282–287
Ishigami A, Asaga H, Ohsawa T et al (2001) Peptidylarginine deiminase type I, type II, type III and type IV are expressed in rat epidermis. Biomed Res 22:63–65
Jang SI, Steinert PM (2002) Loricrin expression in cultured human keratinocytes is controlled by a complex interplay between transcription factors of the Sp1, CREB, AP1, and AP2 families. J Biol Chem 277:42268–42279
Kamata Y, Taniguchi A, Yamamoto M et al (2009) Neutral cysteine protease bleomycin hydrolase is essential for the breakdown of deiminated filaggrin into amino acids. J Biol Chem 284:12829–12836
Kamata Y, Yamamoto M, Kawakami F et al (2011) Bleomycin hydrolase is regulated biphasically in a differentiation- and cytokine-dependent manner: relevance to atopic dermatitis. J Biol Chem 286:8204–8212
Kim JK, Mastronardi FG, Wood DD et al (2003) Multiple sclerosis: an important role for post-translational modifications of myelin basic protein in pathogenesis. Mol Cell Proteomics 2:453–462
Kizawa K, Takahara H, Troxler H et al (2008) Specific citrullination causes assembly of a globular S100A3 homotetramer: a putative CA2+-modulator matures human hair cuticle. J Biol Chem 283:5004–5013
Klareskog L, Ronnelid J, Lundberg K et al (2008) Immunity to citrullinated proteins in rheumatoid arthritis. Annu Rev Immunol 26:651–675
Klose RJ, Zhang Y (2007) Regulation of histone methylation by demethylimination and demethylation. Nat Rev Mol Cell Biol 8:307–318
Lee JH, Jang SI, Yang JM et al (1996) The proximal promoter of the human transglutaminase 3 gene. Stratified squamous epithelial-specific expression in cultured cells is mediated by binding of Sp1 and ets transcription factors to a proximal promoter element. J Biol Chem 271:4561–4568
Li Q, Peterson KR, Fang X, Stamatoyannopoulos G (2002) Locus control regions. Blood 100:3077–3086
Li P, Yao H, Zhang Z et al (2008) Regulation of p53 target gene expression by peptidylarginine deiminase 4. Mol Cell Biol 28:4745–4758
Li P, Li M, Lindberg MR et al (2010) PAD4 is essential for antibacterial innate immunity mediated by neutrophil extracellular traps. J Exp Med 207:1853–1862
Madison KC (2003) Barrier function of the skin: “la raison d'être” of the epidermis. J Invest Dermatol 121:23–41
Markova NG, Karaman-Jurukovska N, Pinkas-Sarafova A et al (2007) Inhibition of histone deacetylation promotes abnormal epidermal differentiation and specifically suppresses the expression of the late differentiation marker profilaggrin. J Invest Dermatol 127:1126–1139
Martin N, Patel S, Segre JA (2004) Long-range comparison of human and mouse Sprr loci to identify conserved noncoding sequences involved in coordinate regulation. Genome Res 14:2430–2438
Méchin M-C, Enji M, Nachat R et al (2005) The peptidylarginine deiminases expressed in human epidermis differ in their substrate specificities and subcellular locations. Cell Mol Life Sci 62:1984–1995
Méchin M-C, Coudane F, Adoue V et al (2010) Deimination is regulated at multiple levels including auto-deimination of peptidylarginine deiminases. Cell Mol Life Sci 67:1491–1503
Nachat R, Méchin M-C, Takahara H et al (2005a) Peptidylarginine deiminase isoforms 1-3 are expressed in the epidermis and involved in the deimination of K1 and filaggrin. J Invest Dermatol 124:384–393
Nachat R, Méchin M-C, Charveron M et al (2005b) Peptidylarginine deiminase isoforms are differentially expressed in the anagen hair follicles and other human skin appendages. J Invest Dermatol 125:34–41
O’Keefe EJ, Hamilton EH, Lee SC, Steinert P (1993) Trichohyalin: a structural protein of hair, tongue, nail, and epidermis. J Invest Dermatol 101(1 Suppl):65S–71S
Palmer CN, Irvine AD, Terron-Kwiatkowski A et al (2006) Common loss-of-function variants of the epidermal barrier protein filaggrin are a major predisposing factor for atopic dermatitis. Nat Genet 38:441–446
Pálmer HG, Anjos-Afonso F, Carmeliet G et al (2008) The vitamin D receptor is a Wnt effector that controls hair follicle differentiation and specifies tumor type in adult epidermis. PLoS One 3:e1483
Rawlings AV, Matts PJ (2005) Stratum corneum moisturization at the molecular level: an update in relation to the dry skin cycle. J Invest Dermatol 124:1099–1110
Rogers GE, Simmonds DH (1958) Content of citrulline and other amino-acids in a protein of hair follicles. Nature 182:186–187
Rothnagel JA, Rogers GE (1986) Trichohyalin, an intermediate filament-associated protein of the hair follicle. J Cell Biol 102:1419–1429
Sandilands A, Terron-Kwiatkowski A, Hull PR et al (2007) Comprehensive analysis of the gene encoding filaggrin uncovers prevalent and rare mutations in ichthyosis vulgaris and atopic eczema. Nat Genet 39:650–654
Sebbag M, Chapuy-Regaud S, Auger I et al (2004) Clinical and pathophysiological significance of the autoimmune response to citrullinated proteins in rheumatoid arthritis. Joint Bone Spine 71:493–502
Senshu T, Kan S, Ogawa H et al (1996) Preferential deimination of keratin K1 and filaggrin during the terminal differentiation of human epidermis. Biochem Biophys Res Commun 225:712–719
Slack JL, Causey CP, Thompson PR (2011a) Protein arginine deiminase 4: a target for an epigenetic cancer therapy. Cell Mol Life Sci 68:709–720
Slack JL, Jones LE Jr, Bhatia MM, Thompson PR (2011b) Autodeimination of protein arginine deiminase 4 alters protein-protein interactions but not activity. Biochemistry 50:3997–4010
Smith FJ, Irvine AD, Terron-Kwiatkowski A et al (2006) Loss-of-function mutations in the gene encoding filaggrin cause ichthyosis vulgaris. Nat Genet 38:337–342
Stacey SN, Gudbjartsson DF, Sulem P et al (2008) Common variants on 1p36 and 1q42 are associated with cutaneous basal cell carcinoma but not with melanoma or pigmentation traits. Nat Genet 40:1313–1318
Steinert PM, Parry DA, Marekov LN (2003) Trichohyalin mechanically strengthens the hair follicle: multiple cross-bridging roles in the inner root sheath. J Biol Chem 278:41409–41419
Struyf S, Noppen S, Loos T et al (2009) Citrullination of CXCL12 differentially reduces CXCR4 and CXCR7 binding with loss of inflammatory and anti-HIV-1 activity via CXCR4. J Immunol 182:666–674
Tarcsa E, Marekov LN, Mei G et al (1996) Protein unfolding by peptidylarginine deiminase. Substrate specificity and structural relationships of the natural substrates trichohyalin and filaggrin. J Biol Chem 271:30709–30716
Tarcsa E, Marekov LN, Andreoli J et al (1997) The fate of trichohyalin. Sequential post-translational modifications by peptidyl-arginine deiminase and transglutaminases. J Biol Chem 272:27893–27901
Urano Y, Watanabe K, Sakaki A et al (1990) Immunohistochemical demonstration of peptidylarginine deiminase in human sweat glands. Am J Dermatopathol 12:249–255
Vossenaar ER, Zendman AJ, van Venrooij WJ et al (2003) PAD, a growing family of citrullinating enzymes: genes, features and involvement in disease. Bioessays 25:1106–1118
Vossenaar ER, Radstake TR, van der Heijden A et al (2004) Expression and activity of citrullinating peptidylarginine deiminase enzymes in monocytes and macrophages. Ann Rheum Dis 63:373–381
Wong CF, Barnes LM, Dahler AL et al (2005) E2F suppression and Sp1 overexpression are sufficient to induce the differentiation-specific marker, transglutaminase type 1, in a squamous cell carcinoma cell line. Oncogene 24:3525–3534
Yamazaki M, Ishidoh K, Suga Y et al (1997) Cytoplasmic processing of human profilaggrin by active mu-calpain. Biochem Biophys Res Commun 235:652–656
Yao H, Li P, Venters BJ et al (2008) Histone Arg modifications and p53 regulate the expression of OKL38, a mediator of apoptosis. J Biol Chem 283:20060–20068
Ying S, Kojima T, Kawada A et al (2010) An intronic enhancer driven by NF-κB contributes to transcriptional regulation of peptidylarginine deiminase type I gene in human keratinocytes. J Invest Dermatol 130:2543–2552
Acknowledgements
We are indebted to all members of HT and GS laboratories involved in PAD research in the past and/or in the present time. HT laboratory is supported by Grants-in-Aid for Scientific Research from the Ministry of Education, Science, Sports, and Culture of Japan. Investigation of PADs in the GS laboratory has been or is supported by the CNRS, the University of Toulouse III, the INSERM, the French Society for Dermatology (SFD), the Society for Dermatological Research (SRD) and Pierre Fabre Dermo-Cosmétique.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Takahara, H., Serre, G., Simon, M. (2014). Deimination in Skin and Regulation of Peptidylarginine Deiminase Expression in Keratinocytes. In: Nicholas, A., Bhattacharya, S. (eds) Protein Deimination in Human Health and Disease. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-8317-5_7
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8317-5_7
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-8316-8
Online ISBN: 978-1-4614-8317-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)