Abstract
Death ligands, a subset of the tumour necrosis factor (TNF) super family, play an important role in innate immunity, inflammatory disorders and cancer and have been the focus of intense study over the past two decades. Depending on the cellular context, the binding of these ligands to their cognate receptors (death receptors) can mediate different cell outcomes: gene activation and cell death. Whilst death receptor-mediated apoptosis has received most of the attention over the years, only recently TNF-induced necrosis has entered the spotlight. Although necrosis was long regarded as an unregulated form of cell death, death ligand-induced necrosis—also referred to as necroptosis—has recently been demonstrated to be dependent on the activity of the kinase activities of the receptor-interacting proteins 1 and 3 (RIP1 and RIP3) and, hence, to be a regulated process. This discovery revealed the opportunity of pharmacological interference with this form of cell death which appears to be involved in a number of pathological conditions. Signalling downstream of TNF is heavily dependent on degradative and non-degradative ubiquitination events. Here, we discuss the current understanding of how the necroptosis pathway is regulated and how the ubiquitin system is thought to control the balance between gene induction, apoptosis and necroptosis.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
5.1 Introduction
Programmed cell death is critically important for the development and survival of multicellular organisms. Hence, deregulation of cell death processes lies at the root of many human diseases including neurodegenerative disorders, immunodeficiencies, autoimmune diseases and cancer. Apoptosis, the best studied form of programmed cell death, relies on signalling pathways leading to the activation of caspases, an evolutionarily conserved family of cysteine proteases. During apoptosis, caspases cleave a large number of substrates in the cell, facilitating efficient removal of the cellular corpse. In contrast, necrosis was long regarded as a form of accidental and unregulated cell death caused by physical stresses. Necrotic cell death occurs independently from caspases and lacks the morphological features that characterise apoptotic cells. However, there is a growing body of evidence that necrotic cell death can also occur in a regulated, programmed manner (Vandenabeele et al. 2010). This mode of cell death is referred to as programmed necrosis or necroptosis. Whereas apoptosis is generally thought of as an anti-inflammatory form of cell death, through the release of damage-associated molecular patterns (DAMPs), necrosis is thought to result in inflammation.
The first findings that indicated that necrosis can occur as a regulated process came from research on members of the tumour necrosis factor (TNF) superfamily (TNFSF) of cytokines. TNF, the first identified member of this family, is a pleiotropic cytokine involved in inflammatory responses, immunity and cell death (Balkwill 2009; Walczak 2011). TNFSF cytokines bind to and cross-link their cognate receptors, the members of the TNF-receptor superfamily (TNF-RSF). These single-pass membrane-spanning receptors are characterised by the presence of cysteine-rich domains in their extracellular portion. Death receptors (DR) constitute a subset of the TNF-RSF and are characterised by the presence of an intracellular death domain (DD). The DR subfamily includes TNF-R1, TRAMP (DR3), CD95 (Fas/APO-1), TRAIL-R1 (DR4) and TRAIL-R2 (KILLER/TRICK/DR5/Apo2) as bona fide members and DR6 as a less well-characterised member (Walczak 2013). DRs are able to mediate programmed cell death by apoptosis following ligation by their respective ligands, at least under certain conditions. The ligands of the bona fide members are TNF, TL1A, CD95L (FasL/APO-1L) and TNF-related apoptosis inducing ligand (TRAIL, also known as Apo2L), respectively. The signalling mechanisms induced by TNF, CD95L and TRAIL are best characterised. Whereas TNF can mediate its effects through binding to two different receptors, TNF-R1 and TNF-R2, of which only TNF-R1 contains a DD (Gray et al. 1984; Loetscher et al. 1990; Pennica et al. 1984; Schall et al. 1990; Smith et al. 1990), CD95L can induce cell death via CD95 (Itoh et al. 1991; Oehm et al. 1992) or be inhibited by the soluble receptor DcR3 (Pitti et al. 1998). TRAIL can induce apoptosis through binding to its DD-containing receptors TRAIL-R1 and TRAIL-R2 (Pan et al. 1997a, b; Screaton et al. 1997; Sheridan et al. 1997; Walczak et al. 1997; Wu et al. 1997). However, TRAIL can also bind to three other receptors, TRAIL-R3, TRAIL-R4 and osteoprotegerin (OPG), but these receptors lack a DD and, hence, do not trigger apoptosis (Walczak and Krammer 2000).
Apart from their ability to activate signalling pathways that result in apoptosis, DRs are also capable of gene activation and induction of pro-inflammatory cytokines. The intracellular signalling pathways involved include the NF-κB pathway and MAP kinase pathways (ERK and JNK pathways). Although the precise mechanisms are currently unclear, the relative balance between the pro-apoptotic and gene activation signals may be determined by several factors, including cell type, strength of ligand-induced signal and presence or absence of certain intracellular proteins that form part of the signalling complexes triggered by the different ligands. When the gene activation pathway dominates over the pro-apoptotic pathway, the consequence would be resistance to ligand-induced cell death.
TNF has long been known to also induce necrosis. In fact, when TNF was first identified, it was shown to selectively induce necrosis in the mouse fibrosarcoma cell line L929 (Carswell et al. 1975). Intriguingly, inhibition of caspases, the central executioners of apoptotic cell death, was found to exacerbate the necrosis-inducing capacity of TNF, and this observation has been extended to cell death induction by TRAIL and CD95L (Holler et al. 2000; Vercammen et al. 1998a, b). Hence, TNF, TRAIL and CD95L are not only able to induce apoptosis but also necrosis.
So why was the necrosis-inducing capacity of TNF and its siblings virtually ignored by the cell death research community during the past two decades? The reason for this is quite simple: apoptosis took centre stage and pushed necrosis off into a “Schattendasein”. During this time there were some scattered reports about TNF-, TRAIL- or CD95L-induced non-apoptotic cell death being necrotic, and receptor-interacting protein 1 (RIP1) was the most likely candidate to be involved in mediating this (Holler et al. 2000). However, it was not until 2008 that Degterev et al. discovered that TNF-induced necrosis can in fact be prevented pharmacologically, indeed by interfering with the kinase activity of RIP1 (Degterev et al. 2008). This discovery has reignited interest in studying DR-mediated necrosis induction because it suddenly appeared feasible to pharmacologically interfere with this type of cell death, i.e. a cell death that could not only not be inhibited by caspase inhibitors but was even exacerbated by caspase inhibition. Much hope was suddenly placed on specific inhibitors of this newly discovered form of programmed necrosis, also referred to as necroptosis, in regard to the possible treatment of several pathological conditions that involve cell death. Clearly, the feat now is to identify the components of and best pharmacological targets within the necroptosis signalling pathways, as well as the matching pathological conditions in which inhibition of programmed necrosis is likely to be beneficial. It is therefore important to gain a detailed understanding of the processes that lead to the triggering of necrosis, both in physiological situations and under pathological conditions, and the exact biochemical mechanisms of necroptosis induction.
Ubiquitination, i.e. the post-translation modification of target proteins by ubiquitin, is a central theme in DR signalling (Bianchi and Meier 2009; Walczak 2011; Wertz and Dixit 2010). In this chapter, we will focus on the role of ubiquitination and its reversal, de-ubiquitination, in DR-mediated signalling that results in necroptosis. Following an introduction into the ubiquitin system and an overview of DR signalling pathways, we will discuss how signalling towards necroptosis is mediated by DRs, and how ubiquitination and de-ubiquitination events in DR-triggered signalling complexes are currently thought to determine the balance between gene activation, apoptosis or necroptosis as outputs of DR triggering.
5.2 The Ubiquitin System
Ubiquitin is a small 8 kDa protein belonging to the family of ubiquitin-like (Ubl) proteins. Ubiquitination is important in a wide range of intracellular signalling processes and normally results in the covalent attachment of ubiquitin to lysine (K) residues of target proteins (Komander and Rape 2012). Ubiquitination involves the action of a three-step enzymatic cascade involving a ubiquitin-activating enzyme (E1), a ubiquitin-conjugating enzyme (E2) and a ubiquitin ligase (E3). In the first step, ubiquitin is activated in an ATP-dependent manner by forming a thioester bond with a cysteine residue in the E1 enzyme. Next, the ubiquitin is transferred from the E1 to the E2 enzyme, which can bind E3s. Finally, the E3, which recruits specific substrates, mediates the transfer of ubiquitin, usually via an isopeptide bond to the lysine residues of the target protein. Ubiquitin E3s can be divided into HECT- and RING-type E3s. In the case of HECT E3 ligases, ubiquitin is first transferred from the E2 to a catalytic cysteine residue of the HECT domain. This ubiquitin-thioester intermediate subsequently transfers ubiquitin to the substrate that is bound to the HECT E3. In contrast to HECT E3s, RING-domain E3s do not form a ubiquitin-thioester intermediate and instead stimulate the direct transfer of ubiquitin from E2 to substrate. An exception to this are the RING E3s that harbour RING-in-between-RING domains (RBR) which were recently shown to function like RING/HECT hybrids whereby the first RING domain binds the E2 after which the ubiquitin moiety is first transferred to a cysteine in the second “RING” domain—which therefore acts as a HECT rather than a RING domain—before ubiquitination of the substrate (Stieglitz et al. 2012; Wenzel et al. 2011). The removal of ubiquitin moieties from protein substrates is catalysed by a diverse class of proteases termed de-ubiquitinases (DUBs).
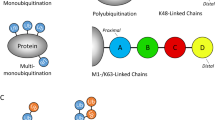
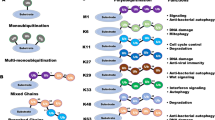
When a single moiety of ubiquitin is covalently attached to a lysine residue of a target protein, this is referred to as mono-ubiquitination. Ubiquitin itself contains seven lysine residues (K6, K11, K27, K29, K33, K48 and K63), and these can in turn be conjugated with another ubiquitin molecule, allowing the formation of ubiquitin chains. Additionally, also the N-terminal methionine (M1) of ubiquitin can be conjugated with another ubiquitin, resulting in M1-linked chains (also called linear chains) (Kirisako et al. 2006; Rieser et al. 2013; Walczak 2011). All lysines and M1 of ubiquitin are present in different positions on the surface of the protein. Therefore, depending on which ubiquitin-contained lysine or whether M1 is used for ubiquitin chain formation, each type of ubiquitin linkage adopts a structurally distinct conformation (Komander 2009). The different ubiquitin linkages are connected to specific downstream signalling processes which are, in turn, translated into distinct cellular functions induced by specific proteins that are termed ubiquitin receptors. Ubiquitin receptors harbour ubiquitin-binding domains (UBDs) that allow them to recognise the different ubiquitin modifications through non-covalent interactions, in many cases in a linkage-type-specific manner (Dikic et al. 2009). Ubiquitination-dependent signalling is terminated by the action of DUBs that specifically hydrolyse ubiquitin chains, sometimes in a linkage-specific manner, and thereby counteract the action of E3 ligases.
5.3 Death Receptor Signalling
Two groups of DRs can be identified: those that recruit the adaptor protein Fas-associated death domain (FADD) (CD95, TRAIL-R1, TRAIL-R2) and those that recruit the adaptor protein TNFR-associated death domain (TRADD) (TNF-R1 and DR3). The primary signalling outcome for the FADD-recruiting receptors is cell death, whilst recruitment of TRADD results in gene activation as the primary signalling output (Fig. 5.1). Binding of TRAIL to TRAIL-R1 and/or TRAIL-R2, and of CD95L to CD95, results in the formation of the death-inducing signalling complex (DISC). First, the adaptor protein FADD is recruited to the intracellular portion of the receptor(s) whose cross-linking was triggered by the respective ligand. This happens through homotypic interaction between the DD of FADD and the DD of TRAIL-R1/R2 or that of CD95. In addition to a DD, FADD also contains a death effector domain (DED) through which it recruits the two initiator caspases, caspase-8 and caspase-10. Recruitment of caspase-8/10 to FADD results in their activation at the DISC. Active caspase-8/10 subsequently undergoes a series of autocatalytic events, which separates the pro-domain from the caspase domains, and stabilises caspase-8/10 dimers. Subsequently, fully activated caspase-8 cleaves the downstream effector caspases caspase-3 and caspase-7, which results in apoptotic cell death.
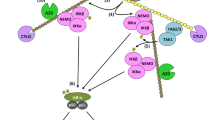
Death receptor signalling. TNF-R1 signalling (left) and TRAIL-R1, TRAIL-R2 and CD95 signalling (right) are depicted. DRs trigger two main signals. The primary signalling output for TNF-R1 is gene activation via NF-κB and MAPK, whereas CD95, TRAIL-R1 and TRAIL-R2 primarily induce apoptosis. For all DR systems, the protein complex that forms at the plasma membrane and exerts the primary function of the respective receptor is defined as complex-I. Binding of TNF to its receptor TNF-R1 results in the formation of the TNF-R1 signalling complex (TNF-RSC). Cross-linked TNF-R1 recruits RIP1 and the adaptor protein TRADD to the DD of the receptor. Subsequently, TRADD recruits TRAF2, which in turn provides the platform for recruitment of the ubiquitin E3 ligases cIAP1 and cIAP2. cIAP1/2 then place various ubiquitin chains of different linkages on several components of the TNF-RSC. cIAP1/2-mediated ubiquitination of TNF-RSC components allows recruitment of LUBAC. Once recruited, LUBAC places M1-linked (linear) ubiquitin chains on RIP1 and NEMO. The different ubiquitin chains generated by cIAP1/2 and LUBAC in the TNF-RSC enable recruitment of the IKK and TAB/TAK complexes, which leads to downstream gene activation. Binding of TRAIL or CD95L to their cognate receptors leads to the formation of the death-inducing signalling complex (DISC). First, the adaptor protein FADD is recruited by a homotypic interaction of the DD of one FADD molecule with the DDs of three ligand-cross-linked receptors. Subsequently, the pro-forms of caspase-8 and caspase-10 are recruited by interaction of their DED with that of FADD. Oligomerisation of caspase-8/10 in the DISC leads to its activation and induction of apoptosis. FLIP isoforms can also be recruited to the DISC, resulting in inhibition of apoptosis by completely or partially preventing caspase-8 activation. The primary complexes can dissociate from the DD of the respective receptor and recruit additional proteins from the cytosol to form a secondary complex, also called complex-II, which triggers the respective secondary signal. In the case of TNF-R1, this can be the induction of necroptosis or apoptosis (see Fig. 5.2), whereas in the case of CD95 and TRAIL-R1/2, the secondary signal is gene activation
Caspase-8-mediated apoptosis is negatively regulated at the DISC by the caspase-8-like molecule FLICE-like inhibitory protein (FLIP). FLIP can also be recruited to the DISC through binding to the DED of FADD. The initially discovered form of FLIP was a viral protein that was found to inhibit caspase-8 (Thome et al. 1997). Shortly thereafter, the cellular version of this protein was identified (also called cellular FLIP, cFLIP) (Irmler et al. 1997). In human cancer cells, two FLIP isoforms are prominently expressed: the long, 55 kDa (FLIPL), and the short, 26 kDa (FLIPS), isoforms (Budd et al. 2006). FLIPL resembles caspase-8 in its domain architecture but lacks a catalytic cysteine in the caspase-like domain necessary for enzymatic activity. Hence, FLIPL represents an inactive pseudo-caspase. FLIPS is highly homologous to viral FLIP and only encodes the pro-domain, which contains the DED domains necessary for FADD binding. Dimerisation of FLIPL/S with caspase-8 inhibits full caspase-8 activation and apoptosis. However, the FLIPS and FLIPL isoforms differentially regulate DR-induced necroptosis, as will be discussed in Sect. 5.4. Caspase-8 activation at the DISC can also be regulated by ubiquitination events. For instance, in DR signalling the E3 ligase Cullin-3 interacts with caspase-8 in the DISC and modifies it with K48- and K63-linked poly-ubiquitin chains. The ubiquitin receptor p62 is then recruited to poly-ubiquitinated caspase-8, which promotes its aggregation and activation (Jin et al. 2009). In addition, K48-linked poly-ubiquitination of caspase-8 was also reported to promote its degradation, thereby shutting off apoptotic signalling (Gonzalvez et al. 2012).
Unlike stimulation of the FADD-recruiting receptors, binding of TNF to TNF-R1 does not usually result in cell death but instead induces transcription of genes important for inflammation and cell survival. The TNF-induced signalling pathway that results in gene activation has been best described for the activation of NF-κB transcription factors. NF-κB family members (p65/RelA, c-REL, REL-B, NF- κB1 and NF-κB2) exist as homo- and heterodimers and regulate genes involved in inflammation and cell survival. NF-κB dimers can shuttle between nucleus and cytoplasm, but under resting conditions they are sequestered in the cytoplasm because they are bound to inhibitor of κB (IκB) proteins. Ligation of TNF-R1 by TNF results in the formation of a signalling complex that is referred to as the TNF-R1 signalling complex (TNF-RSC) or complex-I of TNF-R1 signalling (Micheau and Tschopp 2003) (Fig. 5.1). Both TRADD and RIP1 are independently recruited to the DD of TNF-R1 through their respective DDs. TRADD serves as a platform for the recruitment of TRAF2 and/or TRAF5. Whilst TRAF2 carries a RING domain, the role of TRAF2 as an E3 ligase in TNF signalling is controversial (Alvarez et al. 2010; Yin et al. 2009). Although recruitment of TRAF2 to complex-I is required for downstream signalling and gene activation, the RING of TRAF2 is reportedly dispensable for TNF-mediated activation of NF-κB (Vince et al. 2009; Zhang et al. 2010). The importance of TRAF2 for TNF signalling can therefore be attributed to the fact that it serves as a platform for recruitment of the RING E3 ligases cIAP1 and cIAP2 to the TNF-RSC through its cIAP interaction motif (CIM) (Vince et al. 2009). The homologous proteins cIAP1 and cIAP2 are members of the inhibitor of apoptosis (IAP) protein family. It appears that they can, at least in part, compensate for the loss of each other. Nevertheless, subtle functional differences seem to exist between cIAP1 and cIAP2 (Darding et al. 2011; Feltham et al. 2011; Gyrd-Hansen et al. 2008) and it will be interesting to dissect this further. Members of the IAP family, such as XIAP in mammals, are classically known to inhibit apoptotic cell death by binding and inhibiting caspases directly. However, cIAPs do not inhibit caspases directly but instead are implicated in ubiquitin-dependent signalling pathways. Structural studies have demonstrated that the TRAF2-cIAP2 complex consists of a TRAF2 trimer binding to one cIAP2 molecule (Mace et al. 2010; Zheng et al. 2010). TRAF5 is also a RING-containing protein, yet it apparently cannot bind cIAPs which leaves its role in TNF signalling less clear than originally thought (Silke 2011). Following recruitment of cIAP1/2 to the TNF-RSC, cIAPs ubiquitinate various components of complex-I, most prominently RIP1 and cIAP1 itself, with K63- and K11-linked poly-ubiquitin chains (Bertrand et al. 2008; Dynek et al. 2010; Gerlach et al. 2011; Varfolomeev et al. 2008). cIAP1-mediated ubiquitination is critically important for TNF signalling as genetic or pharmacological depletion of cIAPs results in loss of ubiquitination of RIP1 and failure to activate NF-κB (Bertrand et al. 2008; Vince et al. 2007). cIAP1/2-generated ubiquitin linkages, placed on components of complex-I, allow the subsequent recruitment of the linear ubiquitin chain assembly complex (LUBAC) (Gerlach et al. 2011; Haas et al. 2009; Ikeda et al. 2011; Tokunaga et al. 2009, 2011). LUBAC is an E3 ligase complex that consists of two RBR proteins, HOIP (RNF31) and HOIL-1 (RBCK1), and one additional factor, SHARPIN (SIPL1). Currently, LUBAC is the only E3 ligase known to generate M1-ubiquitin linkages under physiological conditions. Moreover, LUBAC exclusively generates linear ubiquitin chains. HOIP is the central component of this complex as the formation of linear linkages depends on the enzymatic activity of the RBR of HOIP (Haas et al. 2009; Smit et al. 2012; Tokunaga et al. 2009). LUBAC is recruited to cIAP1/2-generated linkages by a UBD (a NZF2 domain) present in HOIP. Although cIAPs also contain a UBD, its role as a ubiquitin receptor in TNF signalling is not known (Gyrd-Hansen et al. 2008).
The ubiquitin linkages placed on RIP1—and possibly other components of the TNF-RSC—by cIAPs and LUBAC allow for more efficient recruitment and retention of the kinase complexes consisting of TAK1 and TAB2 or TAB3 and of NEMO, IKKα and IKKβ, i.e. the TAK/TAB and IκB kinase (IKK) complexes, respectively. In the native TNF-RSC, RIP1 is simultaneously modified with K11-, K48-, K63- and M1-linked chains (Gerlach et al. 2011). Recruitment of the TAK/TAB and IKK complexes is mediated by the UBDs present in TAB2 and NEMO, respectively. The ubiquitin binding in ABIN and NEMO (UBAN) domain of NEMO is able to bind to K63-, K11- and M1-linked ubiquitin chains, all of which are present in the native TNF-RSC (Dynek et al. 2010; Gerlach et al. 2011; Lo et al. 2009; Rahighi et al. 2009). However, the affinity of NEMO’s UBAN for M1-linked ubiquitins is 100-fold greater than for K63- and K11-linked ubiquitins. The UBD of TAB2, in turn, does not bind M1-linkages but specifically interacts with K63-linked ubiquitins (Komander et al. 2009). In addition to RIP1, LUBAC modifies NEMO with linear ubiquitin chains. In contrast to RIP1, on NEMO exclusively linear ubiquitin chains have been identified when present in the native TNF-RSC, although it is currently unclear what the functional role of this modification is (Gerlach et al. 2011). Following activation, IKKβ phosphorylates IκB which leads to recognition by the Skp1-Cullin-F-box (SCF)β-TRCP E3 ubiquitin ligase complex. SCFβ-TRCP ubiquitinates IκB with K48-linked chains, thereby targeting it for proteasomal degradation. This liberates NF-κB dimers from IκB-imposed inhibition so that they can now translocate to the nucleus and drive expression of NF-κB target genes.
TNF-induced NF-κB signalling is negatively regulated by its gene products. For instance, IκB is rapidly transcriptionally upregulated upon NF-κB activation. In addition, DUBs such as A20 and Cezanne are upregulated by NF-κB and mediate disassembly of complex-I by hydrolysing the ubiquitin modifications present in the complex. Other DUBs such as CYLD and USP21, whose expression does not dependent on NF-κB, are also implicated in negatively regulating TNF signalling. Although the specific roles of the individual DUBs in the disassembly of the TNF-RSC are not clear, together they achieve removal of the different ubiquitin linkages from the various components of the complex. Notably, RIP1 is usually almost completely stripped of its many ubiquitinations. It is likely that different DUBs are recruited to different ubiquitin linkages on different components of the complex, mediated by their own UBDs or UBDs present in their adaptor proteins. For instance, ABINs are adaptor proteins required for the recruitment of A20 to the TNF-RSC. ABINs contain a UBAN domain, allowing them to bind linear ubiquitin chains (Wagner et al. 2008).
Apart from being recruited to specific ubiquitin linkages, different DUBs appear to be specific in cleaving certain ubiquitin-linkage types. For instance, A20 has been reported to cleave K63-linked chains from RIP1, and subsequently targeting it for degradation, thereby terminating TNF signalling (Wertz et al. 2004). CYLD is able to cleave K63- and M1-linked ubiquitin chains whereas Cezanne reportedly specifically hydrolyses K11-linkages (Bremm et al. 2010; Komander et al. 2009). The targeted recruitment of specific DUBs to certain linkages and their preferences for catalysing specific linkage types probably serve to precisely fine-tune the extent and duration of a given signal.
Whilst gene activation is the primary signalling outcome of TNF stimulation, under certain circumstances TNF can also induce cell death. TNF-induced cell death is mediated by a secondary complex that forms approximately 2 h after TNF stimulation (Micheau and Tschopp 2003). This cytoplasmic complex, which derives from complex-I, is frequently referred to as complex-II. Complex-II forms when TRADD dissociates from the receptor and instead binds to FADD in the cytosol. FADD in turn can serve as a platform for the recruitment of caspase-8 and FLIP. When TNF successfully stimulates NF-κB-mediated expression of target genes, formation of complex-II does not lead to apoptosis because anti-apoptotic genes, such as Bcl-2, cIAP2 and FLIP, are transcriptionally upregulated. Therefore, TNF only kills under conditions where expression of NF-κB target genes is blocked, such as upon genetic deletion of NF-κB, expression of a dominant negative form of IκB or in the presence of the protein synthesis inhibitor cycloheximide. In these cases, FLIP is not upregulated after TNF stimulation, allowing full caspase-8 activation in complex-II and induction of apoptosis. As will be discussed further in the next sections of this chapter, complex-II can also be formed when ubiquitination events in complex-I are perturbed, thereby altering the ubiquitination status of RIP1. Unmodified RIP1 can then bind to FADD and caspase-8 and stimulate their association, forming complex-II (Wang et al. 2008).
5.4 Necroptosis Signalling
As mentioned above, apart from gene activation and apoptosis, stimulation with death ligands can also lead to the induction of necroptosis, in particular in the presence of caspase inhibitors. As necroptosis has only recently been discovered, at present the necroptosis signalling pathway is not well understood but currently under intense investigation. Importantly, whilst RIP1 plays a central role in ubiquitin-mediated NF-κB signalling downstream of TNF-R1, RIP1 was also the first factor found to be required for death-ligand-induced necroptosis in the presence of caspase inhibition (Holler et al. 2000). Subsequently, the kinase activity of RIP1 was found to be the target of necrostatin-1, a chemical inhibitor of necroptosis (Degterev et al. 2008). In addition to the kinase activity of RIP1, TNF- and virus-induced necroptosis is dependent on the RIP1-related kinase RIP3 (Cho et al. 2009; He et al. 2009; Hitomi et al. 2008; Zhang et al. 2009). Whereas RIP1 is involved in the induction of apoptosis, necrosis and gene activation by TNF, RIP3 appears to play a more specific role as a key determinant of necroptosis. RIP3 and RIP1 are similar in structure as they both harbour an N-terminal kinase domain and can associate with each other through their respective C-terminal RIP homotypic interaction motif (RHIM) domains. However, RIP3 lacks the DD through which RIP1 is capable of interacting with other DD-containing proteins such as FADD, TRADD or TNF-R1 itself.
When caspase activity is inhibited, RIP1 in complex-II is now able to recruit RIP3 to form a pro-necroptotic signalling complex frequently referred to as the necrosome. The kinase activity of RIP1 is required for necrosome formation as this can be inhibited by necrostatin-1 (Cho et al. 2009). Both RIP1 and RIP3 are highly phosphorylated during necroptosis, which again can be inhibited by necrostatin-1, suggesting that RIP1 is the upstream kinase responsible for phosphorylation of its target RIP3 (Cho et al. 2009; Degterev et al. 2008). Phosphorylation of RIP3 on S227 is needed for recruitment of the mixed-lineage kinase-like (MLKL) protein, a pseudo-kinase that was recently identified as a crucial factor for necroptotic signalling downstream of RIP1 and RIP3 (Sun et al. 2012; Zhao et al. 2012). MLKL interacts with phosphorylated RIP3 through its C-terminal kinase-like domain during necroptosis induction. Following recruitment, MLKL is phosphorylated by RIP3 at T357 and S358 (Chen et al. 2013; Sun et al. 2012). MLKL was identified as the target of the small molecule inhibitor of necroptosis necrosulfonamide (NSA) in a chemical library screen (Sun et al. 2012). NSA blocks necroptosis by covalently binding to Cys86 in the N-terminal coiled-coil (CC) domain of MLKL, indicating that this domain is important for MLKL’s function. As this residue is not conserved in murine MLKL, NSA is unable to block necroptosis in mouse cells. One concept that has recently been put forward, but awaits confirmation, is that RIP1/RIP3/MLKL-containing necrosomes aggregate into amyloid-like structures, resulting in the manifestation of the physical features of necroptosis (Li et al. 2012). Although several other components have been implicated in the execution of necroptosis, the biochemical mechanism of necroptosis induction is not yet understood and formal genetic proof for the requirement of any factors for the execution of necroptosis, apart from RIP1, RIP3 and MLKL, is currently lacking.
Recent studies in mice deficient for key components of the apoptosis machinery provided insight on the mechanisms that regulate RIP1-/RIP3-dependent necroptosis (Dillon et al. 2012; Kaiser et al. 2011; Oberst et al. 2011; Zhang et al. 2011) (Fig. 5.2). At the same time, they provided an explanation for the puzzling observation that deficiency in caspase-8, FADD or FLIP results in an embryonically lethal phenotype that cannot be attributed to deregulated apoptosis. Since all of these three gene-deficient mice die at the same stage of embryonic development at around E10.5, displaying a phenotype that showed lack of vascularisation of the yolk sac (Sakamaki et al. 2002; Varfolomeev et al. 1998), it could be deduced that caspase-8, FADD and FLIP likely act in concert to prevent the death of a precursor cell of yolk sac vascularisation. Until recently it was, however, unclear whether this hypothesis was correct and, if so, what type of cell death the caspase-8/FLIP/FADD triad would be capable of inhibiting. Strikingly, normal development is completely restored in caspase-8- and FADD-deficient mice when RIP3 is absent (Kaiser et al. 2011; Oberst et al. 2011). In addition, the embryonic lethality of FADD-deficient mice is also rescued by the deletion of RIP1 (Zhang et al. 2011). However, similar to RIP1-deficient mice, these animals die shortly after birth. These studies revealed that, together, FADD, caspase-8 and FLIP antagonise RIP1-/RIP3-dependent necroptosis signalling.
Necroptosis signalling. The different cell death outcomes downstream of TNF-R1 are depicted. Following formation of the TNF-RSC (complex-I), components of this complex, notably TRADD and RIP1, can be released into the cytosol, forming a secondary complex referred to as complex-II. This process is most likely facilitated by the action of DUBs. Complex-II can recruit FADD, caspase-8/10, FLIP isoforms and RIP3, when expressed. The presence of FLIP isoforms in complex-II inhibits full caspase-8 activation and hence apoptosis. The caspase-8/FLIPL heterodimer also counteracts RIP1 and RIP3 signalling, thereby preventing necroptosis. When FLIP levels are low, for example, when the gene activation signals of complex-I are inhibited, caspase-8 activation is uncontrolled and leads to apoptosis. When caspase-8 activity is completely absent, for instance, by genetic deletion or by the presence of high levels of FLIPS in complex-II, RIP1 and RIP3 signalling is unchecked. The kinase activities of RIP1 and RIP3 allow their stable oligomerisation and recruitment and phosphorylation of MLKL, resulting in execution of necroptosis by a yet undefined downstream pathway
What could be the explanation for these observations on the molecular level? The different FLIP isoforms appear to be the key determinants in deciding cell fate. Whilst FLIPS completely inhibits caspase-8 activation, counter-intuitively FLIPL actually appears to promote localised activation of caspase-8 (Boatright et al. 2004; Micheau et al. 2002; Oberst et al. 2011; Pop et al. 2011) (Fig. 5.2). FLIPL-mediated activation of caspase-8 is achieved through dimerisation-induced conformational changes of the FLIPL-caspase-8 heterodimer. Heterodimerisation of FLIPL and caspase-8 allows cleavage between their respective large and small subunits and, consequently, activation of the caspase-8 portion of the heterodimer. Although FLIPL thereby imparts catalytic activity to caspase-8, this activity is not sufficient to trigger cell death, because FLIPL prevents cleavage of the pro-domain of caspase-8. Moreover, it appears that FLIPL-caspase-8 heterodimers display less proteolytic activity than caspase-8 homodimers on apoptotic substrates such as Bid and caspase-3 (Hughes et al. 2007; Pop et al. 2011). FADD, serving as the scaffold for caspase-8 and FLIPL, is also able to bind RIP1. The current model is that the FADD/caspase-8/FLIPL complex allows localised caspase-8 activity that results in cleavage of RIP1 and RIP3 which, in turn, probably results in their inactivation (Feng et al. 2007; Oberst et al. 2011). Hence, FLIPL inhibits both caspase-8-mediated apoptosis and RIP1-/RIP3-dependent necroptosis (Fig. 5.2). In contrast, FLIPS, which completely prevents catalytic activity of caspase-8, cannot inhibit RIP1/3-dependent necroptosis. Since FLIPL inhibits both apoptosis and necroptosis, in FLIP knockout mice both pathways need to be neutralised in order to rescue the lethal phenotype. Accordingly, embryonic lethality in mice lacking FLIP cannot be rescued by deletion of RIP3 because in the absence of FLIP, apoptotic cell death can occur in an uncontrolled manner and, indeed, these embryos display apoptotic cell death (Dillon et al. 2012). Consistent with this notion, the deletion of FADD in addition to FLIP and RIP3 resulted in viable FLIP/FADD/RIP3 triple-knockout mice (Dillon et al. 2012). Thus, the components of the apoptotic machinery, FADD, caspase-8 and FLIP have a survival role by means of negatively regulating RIP1 and RIP3, thereby controlling aberrant necroptosis in the developing embryo.
Currently, it is not clear what the physiological roles of necroptosis are. Whereas RIP1-deficient mice die perinatally, both RIP3- and MLKL-deficient mice are healthy and do not display any remarkable developmental defects (Cho et al. 2009; He et al. 2009; Wu et al. 2013). Whilst these knockouts show that necroptosis is not required for normal development, they also provide invaluable means to study the pathophysiological role of necroptosis. Evidence is emerging that necroptosis is likely to play a crucial role in the host defence against viruses and other pathogens. As viruses commonly bear genes expressing apoptosis inhibitors, RIP3-dependent necrosis most likely serves as a second-line defence cell death mechanism to eliminate infected cells when apoptosis is inhibited. Indeed, although RIP3-deficient mice are developmentally normal, they are unable to control viral infections (Cho et al. 2009; Upton et al. 2010). The notion that necroptosis is a crucial component of anti-viral immunity is further supported by the recent finding that certain viruses also express necroptosis inhibitors, such as the cytomegalovirus protein M45/vIRA that binds and inhibits RIP3 (Upton et al. 2010). In addition, it has been suggested that necroptosis of infected cells enhances the inflammatory response since necroptotic cells release pro-inflammatory DAMPs (Weinlich et al. 2011).
5.5 Regulation of Necroptosis by Ubiquitination
As discussed above, ubiquitination events play an important role in TNF signalling. In fact, over the years the ubiquitin system has emerged as a key regulator of cell death and survival signalling. The first indications of the importance of ubiquitination came from studies on apoptotic signalling in Drosophila melanogaster. The drosophila IAPs, DIAP1 and DIAP2, limit caspase activation by modifying both initiator and effector caspases with degradative and non-degradative ubiquitin chains (Broemer and Meier 2009). In mammals, recent evidence suggests that IAPs can regulate caspase-8 activation directly and independently from DR signalling. Loss of IAPs, either due to genotoxic stress or pharmacological inhibition, results in spontaneous formation of a RIP1-containing cytoplasmic death-inducing complex dubbed the “Ripoptosome” (Feoktistova et al. 2011; Tenev et al. 2011). Similar to complex-II, the Ripoptosome contains the core components RIP1, caspase-8, FLIP and FADD and can stimulate caspase-8-dependent apoptosis as well as necroptosis when RIP3 is present. An important difference with complex-II is that the Ripoptosome is assembled in the cytosol independently from DR signalling. Most likely, cIAP1, cIAP2 and XIAP negatively regulate the Ripoptosome by targeting its components for ubiquitination and degradation.
Downstream of TNF-R1, the formation of ubiquitin linkages by cIAP1/2 plays an important role in regulating the signalling outcome. First of all, as outlined above, NF-κB activation downstream of TNF-R1 depends on various degradative and non-degradative ubiquitination events. In addition, ubiquitination of RIP1, and possibly other components of the TNF-RSC, by cIAPs and LUBAC, is important to actively protect cells against the lethal effects of TNF. The importance of IAPs in TNF signalling was manifested by the use of Smac mimetics (SMs). SMs are a class of small molecule drugs that mimic the N-terminal IAP-binding motif (IBM) of the mature form of the endogenous IAP antagonist Smac. Originally, these compounds were designed to cause apoptosis of cancer cells by antagonising XIAP and relieving caspases from its inhibition (Li et al. 2004; Schimmer et al. 2004). However, SM compounds exert their strongest effect on cIAP1 and cIAP2 by causing their proteasomal degradation (Petersen et al. 2007; Varfolomeev et al. 2007; Vince et al. 2007). These two cellular IAPs normally mediate the constitutive degradation of NF-κB-inducing kinase (NIK), a kinase that mediates non-canonical activation of NF-κB. Depletion of cIAPs by treatment with SMs results in stabilisation of NIK and spontaneous activation of non-canonical NF-κB signalling. In some cells, SM-mediated activation of NF-κB leads to induction of TNF, resulting in subsequent autocrine stimulation of TNF-R1. In the absence of cIAPs, cells are sensitised to the lethal effects of TNF since components of complex-I, in particular RIP1, are no longer ubiquitinated. The lack of ubiquitination in complex-I leads to the formation of the cytoplasmic complex-II, which is dependent on RIP1 (Petersen et al. 2007; Wang et al. 2008). RIP1 promotes the association between caspase-8 and FADD in complex-II, resulting in caspase-8 activation and induction of apoptosis (Fig. 5.2). When RIP3 is present, however, it can also be recruited to RIP1 in complex-II; when, in addition, caspase-8 is inhibited, the combination of SM treatment and TNF stimulation can result in necroptosis.
Apart from the E3 ligase activity of cIAPs, linear ubiquitination of components of complex-I by LUBAC is also required to protect cells against the lethal effects of TNF. Interestingly, mice carrying a mutation in the LUBAC component SHARPIN, which results in loss of SHARPIN expression, develop chronic proliferative dermatitis (cpdm) (Seymour et al. 2007). Whilst gene induction by TNF and other inflammatory ligands is attenuated, cpdm cells are sensitised to TNF-induced cell death (Gerlach et al. 2011; Ikeda et al. 2011; Tokunaga et al. 2011). The observation that TNF stimulation results in aberrant death of cpdm-derived cells and that this cell death is both apoptotic and necroptotic—and hence possibly inflammatory—inspired the cross between TNF-deficient and cpdm mice, with the result that the formation of inflammatory lesions that characterises cpdm mice was completely prevented in the absence of TNF (Gerlach et al. 2011). These data strongly support a model according to which TNF-induced cell death, the only TNF signal that was increased in cells derived from SHARPIN-deficient cpdm mice, is causative for inflammation in cpdm mice. It remains to be shown, however, to which extent TNF-induced necroptosis and/or apoptosis contribute to the cpdm-characterising inflammation and whether, together, these two forms of cell death might be required and sufficient for it to occur.
Apart from E3 ligases, DUBs also play an important role in the regulation of DR-mediated necroptosis. CYLD is recruited to the TNF-R1 signalling complex where it targets ubiquitin linkages on RIP1 (Wright et al. 2007). CYLD has been identified in a genome-wide siRNA screen in the mouse cell line L929 to be required for efficient induction of TNF-induced necroptosis (Hitomi et al. 2008). Subsequently, several other studies have confirmed the role of CYLD in necroptosis as well as apoptosis (Bonnet et al. 2011; O’Donnell et al. 2011; Welz et al. 2011). Similar to RIP1 and RIP3, CYLD is also being cleaved by caspase-8 after DR stimulation. This inactivates CYLD and is another mechanism by which caspase-8 suppresses necroptosis (O’Donnell et al. 2011). The finding that CYLD appears to be capable of not only cleaving K63- but also M1-linked ubiquitin chains (Komander et al. 2009) strengthens the notion that linear ubiquitination by LUBAC prevents RIP1 from inducing cell death. Recently, the novel DUB FAM105B/OTULIN/Gumby has been identified as specifically hydrolysing linear ubiquitin linkages, thereby antagonising LUBAC activity (Keusekotten et al. 2013; Rivkin et al. 2013).
Detailed investigations will be required to understand the exact mechanisms by which cIAPs, LUBAC, OTULIN and CYLD, and most likely other E3 ligases and DUBs, regulate the balance between the gene-activatory, apoptotic and necroptotic outcomes of signalling downstream of death receptors. This will not only be important downstream of TNF-R1 but also of TRAIL-R1/2 and CD95, as the necroptotic signalling arm and the role of ubiquitination downstream of these receptors is less well understood. Moreover, the elucidation of hitherto unknown components and regulators of the necroptosis pathway beyond the ubiquitin system will clearly be the focus of future studies. As necroptosis is being reported to be involved in a wider range of pathologies, it will be crucial to completely dissect the signalling pathways that results in necroptosis. Evidently, the understanding of the exact molecular mechanism underlying necroptosis will lead to the identification of novel therapeutic targets and opportunities.
References
Alvarez SE, Harikumar KB, Hait NC, Allegood J, Strub GM, Kim EY, Maceyka M, Jiang H, Luo C, Kordula T et al (2010) Sphingosine-1-phosphate is a missing cofactor for the E3 ubiquitin ligase TRAF2. Nature 465:1084–1088
Balkwill F (2009) Tumour necrosis factor and cancer. Nat Rev Cancer 9:361–371
Bertrand M, Milutinovic S, Dickson K, Ho W, Boudreault A, Durkin J, Gillard J, Jaquith J, Morris S, Barker P (2008) cIAP1 and cIAP2 Facilitate Cancer Cell Survival by Functioning as E3 Ligases that Promote RIP1 Ubiquitination. Mol Cell 30:689–700
Bianchi K, Meier P (2009) A tangled web of ubiquitin chains: breaking news in TNF-R1 signaling. Mol Cell 36:736–742
Boatright KM, Deis C, Denault JB, Sutherlin DP, Salvesen GS (2004) Activation of caspases-8 and −10 by FLIP(L). Biochem J 382:651–657
Bonnet MC, Preukschat D, Welz PS, van Loo G, Ermolaeva MA, Bloch W, Haase I, Pasparakis M (2011) The adaptor protein FADD protects epidermal keratinocytes from necroptosis in vivo and prevents skin inflammation. Immunity 35:572–582
Bremm A, Freund SM, Komander D (2010) Lys11-linked ubiquitin chains adopt compact conformations and are preferentially hydrolyzed by the deubiquitinase Cezanne. Nat Struct Mol Biol 17:939–947
Broemer M, Meier P (2009) Ubiquitin-mediated regulation of apoptosis. Trends Cell Biol 19:130–140
Budd RC, Yeh W-C, Tschopp J (2006) cFLIP regulation of lymphocyte activation and development. Nat Rev Immunol 6:196–204
Carswell EA, Old LJ, Kassel RL, Green S, Fiore N, Williamson B (1975) An endotoxin-induced serum factor that causes necrosis of tumors. Proc Natl Acad Sci U S A 72:3666–3670
Chen W, Zhou Z, Li L, Zhong CQ, Zheng X, Wu X, Zhang Y, Ma H, Huang D, Li W et al (2013) Diverse Sequence Determinants Control Human and Mouse Receptor Interacting Protein 3 (RIP3) and Mixed Lineage Kinase domain-Like (MLKL) Interaction in Necroptotic Signaling. J Biol Chem 288(23):16247–16261
Cho YS, Challa S, Moquin D, Genga R, Ray TD, Guildford M, Chan FK-M (2009) Phosphorylation-driven assembly of the RIP1-RIP3 complex regulates programmed necrosis and virus-induced inflammation. Cell 137:1112–1123
Darding M, Feltham R, Tenev T, Bianchi K, Benetatos C, Silke J, Meier P (2011) Molecular determinants of Smac mimetic induced degradation of cIAP1 and cIAP2. Cell Death Differ 18(8):1376–1386
Degterev A, Hitomi J, Germscheid M, Ch’en IL, Korkina O, Teng X, Abbott D, Cuny GD, Yuan C, Wagner G et al (2008) Identification of RIP1 kinase as a specific cellular target of necrostatins. Nat Chem Biol 4:313–321
Dikic I, Wakatsuki S, Walters KJ (2009) Ubiquitin-binding domains—from structures to functions. Nat Rev Mol Cell Biol 10:659–671
Dillon CP, Oberst A, Weinlich R, Janke LJ, Kang TB, Ben-Moshe T, Mak TW, Wallach D, Green DR (2012) Survival function of the FADD-CASPASE-8-cFLIP(L) complex. Cell Rep 1:401–407
Dynek JN, Goncharov T, Dueber EC, Fedorova AV, Izrael-Tomasevic A, Phu L, Helgason E, Fairbrother WJ, Deshayes K, Kirkpatrick DS et al (2010) c-IAP1 and UbcH5 promote K11-linked polyubiquitination of RIP1 in TNF signalling. EMBO J 29:4198–4209
Feltham R, Bettjeman B, Budhidarmo R, Mace PD, Shirley S, Condon SM, Chunduru SK, McKinlay MA, Vaux DL, Silke J et al (2011) Smac mimetics activate the E3 ligase activity of cIAP1 protein by promoting RING domain dimerization. J Biol Chem 286:17015–17028
Feng S, Yang Y, Mei Y, Ma L, Zhu DE, Hoti N, Castanares M, Wu M (2007) Cleavage of RIP3 inactivates its caspase-independent apoptosis pathway by removal of kinase domain. Cellular Signalling 19:2056–2067
Feoktistova M, Geserick P, Kellert B, Dimitrova DP, Langlais C, Hupe M, Cain K, MacFarlane M, Hacker G, Leverkus M (2011) cIAPs block Ripoptosome formation, a RIP1/caspase-8 containing intracellular cell death complex differentially regulated by cFLIP isoforms. Mol Cell 43:449–463
Gerlach B, Cordier SM, Schmukle AC, Emmerich CH, Rieser E, Haas TL, Webb AI, Rickard JA, Anderton H, Wong WW et al (2011) Linear ubiquitination prevents inflammation and regulates immune signalling. Nature 471:591–596
Gonzalvez F, Lawrence D, Yang B, Yee S, Pitti R, Marsters S, Pham VC, Stephan JP, Lill J, Ashkenazi A (2012) TRAF2 Sets a threshold for extrinsic apoptosis by tagging caspase-8 with a ubiquitin shutoff timer. Mol Cell 48:888–899
Gray PW, Aggarwal BB, Benton CV, Bringman TS, Henzel WJ, Jarrett JA, Leung DW, Moffat B, Ng P, Svedersky LP et al (1984) Cloning and expression of cDNA for human lymphotoxin, a lymphokine with tumour necrosis activity. Nature 312:721–724
Gyrd-Hansen M, Darding M, Miasari M, Santoro MM, Zender L, Xue W, Tenev T, Fonseca PCAD, Zvelebil M, Bujnicki JM et al (2008) IAPs contain an evolutionarily conserved ubiquitin-binding domain that regulates NF-kappaB as well as cell survival and oncogenesis. Nat Cell Biol 10(11):1309–1317
Haas TL, Emmerich CH, Gerlach B, Schmukle AC, Cordier SM, Rieser E, Feltham R, Vince J, Warnken U, Wenger T et al (2009) Recruitment of the linear ubiquitin chain assembly complex stabilizes the TNF-R1 signaling complex and is required for TNF-mediated gene induction. Mol Cell 36:831–844
He S, Wang L, Miao L, Wang T, Du F, Zhao L, Wang X (2009) Receptor interacting protein kinase-3 determines cellular necrotic response to TNF-alpha. Cell 137:1100–1111
Hitomi J, Christofferson D, Ng A, Yao J, Degterev A, Xavier R, Yuan J (2008) Identification of a molecular signaling network that regulates a cellular necrotic cell death pathway. Cell 135:1311–1323
Holler N, Zaru R, Micheau O, Thome M, Attinger A, Valitutti S, Bodmer JL, Schneider P, Seed B, Tschopp J (2000) Fas triggers an alternative, caspase-8-independent cell death pathway using the kinase RIP as effector molecule. Nat Immunol 1:489–495
Hughes DA, Ginsberg L, Baker R, Goodwin S, Milligan A, Richfield L, Mehta AB (2007) Effective treatment of an elderly patient with Gaucher’s disease and Parkinsonism: a case report of 24 months’ oral substrate reduction therapy with miglustat. Parkinson Relat Dis 13:365–368
Ikeda F, Deribe YL, Skanland SS, Stieglitz B, Grabbe C, Franz-Wachtel M, van Wijk SJ, Goswami P, Nagy V, Terzic J et al (2011) SHARPIN forms a linear ubiquitin ligase complex regulating NF-kappaB activity and apoptosis. Nature 471:637–641
Irmler M, Thome M, Hahne M, Schneider P, Hofmann K, Steiner V, Bodmer JL, Schroter M, Burns K, Mattmann C et al (1997) Inhibition of death receptor signals by cellular FLIP. Nature 388:190–195
Itoh N, Yonehara S, Ishii A, Yonehara M, Mizushima S, Sameshima M, Hase A, Seto Y, Nagata S (1991) The polypeptide encoded by the cDNA for human cell surface antigen Fas can mediate apoptosis. Cell 66:233–243
Jin Z, Li Y, Pitti R, Lawrence D, Pham VC, Lill JR, Ashkenazi A (2009) Cullin3-based polyubiquitination and p62-dependent aggregation of caspase-8 mediate extrinsic apoptosis signaling. Cell 137:721–735
Kaiser WJ, Upton JW, Long AB, Livingston-Rosanoff D, Daley-Bauer LP, Hakem R, Caspary T, Mocarski ES (2011) RIP3 mediates the embryonic lethality of caspase-8-deficient mice. Nature 471:368–372
Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, Wauer T, Hospenthal MK, Gyrd-Hansen M, Krappmann D et al (2013) OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell 153:1312–1326
Kirisako T, Kamei K, Murata S, Kato M, Fukumoto H, Kanie M, Sano S, Tokunaga F, Tanaka K, Iwai K (2006) A ubiquitin ligase complex assembles linear polyubiquitin chains. EMBO J 25:4877–4887
Komander D (2009) The emerging complexity of protein ubiquitination. Biochem Soc Trans 37:937–953
Komander D, Rape M (2012) The ubiquitin code. Ann Rev Biochem 81:203–229
Komander D, Reyes-Turcu F, Licchesi JD, Odenwaelder P, Wilkinson KD, Barford D (2009) Molecular discrimination of structurally equivalent Lys 63-linked and linear polyubiquitin chains. EMBO Rep 10:466–473
Li J, McQuade T, Siemer AB, Napetschnig J, Moriwaki K, Hsiao YS, Damko E, Moquin D, Walz T, McDermott A et al (2012) The RIP1/RIP3 necrosome forms a functional amyloid signaling complex required for programmed necrosis. Cell 150:339–350
Li L, Thomas RM, Suzuki H, De Brabander JK, Wang X, Harran PG (2004) A small molecule Smac mimic potentiates TRAIL- and TNFalpha-mediated cell death. Science 305:1471–1474
Lo YC, Lin SC, Rospigliosi CC, Conze DB, Wu CJ, Ashwell JD, Eliezer D, Wu H (2009) Structural basis for recognition of diubiquitins by NEMO. Mol Cell 33:602–615
Loetscher H, Pan YC, Lahm HW, Gentz R, Brockhaus M, Tabuchi H, Lesslauer W (1990) Molecular cloning and expression of the human 55 kd tumor necrosis factor receptor. Cell 61:351–359
Mace PD, Smits C, Vaux DL, Silke J, Day CL (2010) Asymmetric recruitment of cIAPs by TRAF2. J Mol Biol 400:8–15
Micheau O, Thome M, Schneider P, Holler N, Tschopp J, Nicholson DW, Briand C, Grutter MG (2002) The long form of FLIP is an activator of caspase-8 at the Fas death-inducing signaling complex. J Biol Chem 277:45162–45171
Micheau O, Tschopp J (2003) Induction of TNF receptor I-mediated apoptosis via two sequential signaling complexes. Cell 114:181–190
O’Donnell MA, Perez-Jimenez E, Oberst A, Ng A, Massoumi R, Xavier R, Green DR, Ting AT (2011) Caspase 8 inhibits programmed necrosis by processing CYLD. Nat Cell Biol 13:1437–1442
Oberst A, Dillon CP, Weinlich R, McCormick LL, Fitzgerald P, Pop C, Hakem R, Salvesen GS, Green DR (2011) Catalytic activity of the caspase-8-FLIP(L) complex inhibits RIPK3-dependent necrosis. Nature 471:363–367
Oehm A, Behrmann I, Falk W, Pawlita M, Maier G, Klas C, Li-Weber M, Richards S, Dhein J, Trauth BC et al (1992) Purification and molecular cloning of the APO-1 cell surface antigen, a member of the tumor necrosis factor/nerve growth factor receptor superfamily. Sequence identity with the Fas antigen. J Biol Chem 267:10709–10715
Pan G, Ni J, Wei YF, Yu G, Gentz R, Dixit VM (1997a) An antagonist decoy receptor and a death domain-containing receptor for TRAIL. Science 277:815–818
Pan G, O’Rourke K, Chinnaiyan AM, Gentz R, Ebner R, Ni J, Dixit VM (1997b) The receptor for the cytotoxic ligand TRAIL. Science 276:111–113
Pennica D, Nedwin GE, Hayflick JS, Seeburg PH, Derynck R, Palladino MA, Kohr WJ, Aggarwal BB, Goeddel DV (1984) Human tumour necrosis factor: precursor structure, expression and homology to lymphotoxin. Nature 312:724–729
Petersen S, Wang L, Yalcin-Chin A, Li L, Peyton M, Minna J, Harran P, Wang X (2007) Autocrine TNFalpha Signaling Renders Human Cancer Cells Susceptible to Smac-Mimetic-Induced Apoptosis. Cancer Cell 12:445–456
Pitti RM, Marsters SA, Lawrence DA, Roy M, Kischkel FC, Dowd P, Huang A, Donahue CJ, Sherwood SW, Baldwin DT et al (1998) Genomic amplification of a decoy receptor for Fas ligand in lung and colon cancer. Nature 396:699–703
Pop C, Oberst A, Drag M, Van Raam BJ, Riedl SJ, Green DR, Salvesen GS (2011) FLIP(L) induces caspase 8 activity in the absence of interdomain caspase 8 cleavage and alters substrate specificity. Biochem J 433:447–457
Rahighi S, Ikeda F, Kawasaki M, Akutsu M, Suzuki N, Kato R, Kensche T, Uejima T, Bloor S, Komander D et al (2009) Specific recognition of linear ubiquitin chains by NEMO is important for NF-kappaB activation. Cell 136:1098–1109
Rieser E, Cordier SM, Walczak H (2013) Linear ubiquitination: a newly discovered regulator of cell signalling. Trends Biochem Sci 38:94–102
Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, Huang H, Dunham WH, Fukumura R, Xie G et al (2013) The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature 498:318–324
Sakamaki K, Inoue T, Asano M, Sudo K, Kazama H, Sakagami J, Sakata S, Ozaki M, Nakamura S, Toyokuni S et al (2002) Ex vivo whole-embryo culture of caspase-8-deficient embryos normalize their aberrant phenotypes in the developing neural tube and heart. Cell Death Differ 9:1196–1206
Schall TJ, Lewis M, Koller KJ, Lee A, Rice GC, Wong GH, Gatanaga T, Granger GA, Lentz R, Raab H et al (1990) Molecular cloning and expression of a receptor for human tumor necrosis factor. Cell 61:361–370
Schimmer AD, Welsh K, Pinilla C, Wang Z, Krajewska M, Bonneau MJ, Pedersen IM, Kitada S, Scott FL, Bailly-Maitre B et al (2004) Small-molecule antagonists of apoptosis suppressor XIAP exhibit broad antitumor activity. Cancer Cell 5:25–35
Screaton GR, Mongkolsapaya J, Xu XN, Cowper AE, McMichael AJ, Bell JI (1997) TRICK2, a new alternatively spliced receptor that transduces the cytotoxic signal from TRAIL. Curr Biol 7:693–696
Seymour RE, Hasham MG, Cox GA, Shultz LD, Hogenesch H, Roopenian DC, Sundberg JP (2007) Spontaneous mutations in the mouse Sharpin gene result in multiorgan inflammation, immune system dysregulation and dermatitis. Genes Immun 8:416–421
Sheridan JP, Marsters SA, Pitti RM, Gurney A, Skubatch M, Baldwin D, Ramakrishnan L, Gray CL, Baker K, Wood WI et al (1997) Control of TRAIL-induced apoptosis by a family of signaling and decoy receptors. Science 277:818–821
Silke J (2011) The regulation of TNF signalling: what a tangled web we weave. Curr Opin Immunol 23:620–626
Smit JJ, Monteferrario D, Noordermeer SM, van Dijk WJ, van der Reijden BA, Sixma TK (2012) The E3 ligase HOIP specifies linear ubiquitin chain assembly through its RING-IBR-RING domain and the unique LDD extension. EMBO J 31:3833–3844
Smith CA, Davis T, Anderson D, Solam L, Beckmann MP, Jerzy R, Dower SK, Cosman D, Goodwin RG (1990) A receptor for tumor necrosis factor defines an unusual family of cellular and viral proteins. Science 248:1019–1023
Stieglitz B, Morris-Davies AC, Koliopoulos MG, Christodoulou E, Rittinger K (2012) LUBAC synthesizes linear ubiquitin chains via a thioester intermediate. EMBO Rep 13:840–846
Sun L, Wang H, Wang Z, He S, Chen S, Liao D, Wang L, Yan J, Liu W, Lei X et al (2012) Mixed lineage kinase domain-like protein mediates necrosis signaling downstream of RIP3 kinase. Cell 148:213–227
Tenev T, Bianchi K, Darding M, Broemer M, Langlais C, Wallberg F, Zachariou A, Lopez J, MacFarlane M, Cain K et al (2011) The Ripoptosome, a signaling platform that assembles in response to genotoxic stress and loss of IAPs. Mol Cell 43:432–448
Thome M, Schneider P, Hofmann K, Fickenscher H, Meinl E, Neipel F, Mattmann C, Burns K, Bodmer JL, Schroter M et al (1997) Viral FLICE-inhibitory proteins (FLIPs) prevent apoptosis induced by death receptors. Nature 386:517–521
Tokunaga F, Nakagawa T, Nakahara M, Saeki Y, Taniguchi M, Sakata S, Tanaka K, Nakano H, Iwai K (2011) SHARPIN is a component of the NF-kappaB-activating linear ubiquitin chain assembly complex. Nature 471:633–636
Tokunaga F, Sakata S, Saeki Y, Satomi Y, Kirisako T, Kamei K, Nakagawa T, Kato M, Murata S, Yamaoka S et al (2009) Involvement of linear polyubiquitylation of NEMO in NF-kappaB activation. Nat Cell Biol 11(2):123–132
Upton JW, Kaiser WJ, Mocarski ES (2010) Virus inhibition of RIP3-dependent necrosis. Cell Host Microbe 7:302–313
Vandenabeele P, Galluzzi L, Vanden Berghe T, Kroemer G (2010) Molecular mechanisms of necroptosis: an ordered cellular explosion. Nat Rev Mol Cell Biol 11:700–714
Varfolomeev E, Blankenship J, Wayson S, Fedorova A, Kayagaki N, Garg P, Zobel K, Dynek J, Elliott L, Wallweber H et al (2007) IAP antagonists induce autoubiquitination of c-IAPs, NF-kappaB activation, and TNFalpha-dependent apoptosis. Cell 131:669–681
Varfolomeev E, Goncharov T, Fedorova AV, Dynek JN, Zobel K, Deshayes K, Fairbrother WJ, Vucic D (2008) c-IAP1 and c-IAP2 are critical mediators of tumor necrosis factor alpha (TNFalpha)-induced NF-kappaB activation. J Biol Chem 283:24295–24299
Varfolomeev EE, Schuchmann M, Luria V, Chiannilkulchai N, Beckmann JS, Mett IL, Rebrikov D, Brodianski VM, Kemper OC, Kollet O et al (1998) Targeted disruption of the mouse Caspase 8 gene ablates cell death induction by the TNF receptors, Fas/Apo1, and DR3 and is lethal prenatally. Immunity 9:267–276
Vercammen D, Beyaert R, Denecker G, Goossens V, Van Loo G, Declercq W, Grooten J, Fiers W, Vandenabeele P (1998a) Inhibition of caspases increases the sensitivity of L929 cells to necrosis mediated by tumor necrosis factor. J Exp Med 187:1477–1485
Vercammen D, Brouckaert G, Denecker G, Van de Craen M, Declercq W, Fiers W, Vandenabeele P (1998b) Dual signaling of the Fas receptor: initiation of both apoptotic and necrotic cell death pathways. J Exp Med 188:919–930
Vince J, Pantaki D, Feltham R, Mace P, Cordier S, Schmuckle A, Davidson A, Callus B, Wong W, Gentle I et al (2009) TRAF2 must bind to cIAPs for TNF to efficiently activate NF-{kappa}B and to prevent TNF-induced apoptosis. J Biol Chem 284(51):35906–35915
Vince J, Wong W, Khan N, Feltham R, Chau D, Ahmed A, Benetatos C, Chunduru S, Condon S, McKinlay M et al (2007) IAP antagonists target cIAP1 to induce TNFalpha-dependent apoptosis. Cell 131:682–693
Wagner S, Carpentier I, Rogov V, Kreike M, Ikeda F, Lohr F, Wu CJ, Ashwell JD, Dotsch V, Dikic I et al (2008) Ubiquitin binding mediates the NF-kappaB inhibitory potential of ABIN proteins. Oncogene 27:3739–3745
Walczak H (2011) TNF and ubiquitin at the crossroads of gene activation, cell death, inflammation, and cancer. Immunol Rev 244:9–28
Walczak H (2013) Death receptor-ligand systems in cancer, cell death, and inflammation. Cold Spring Harbor Perspect Biol 5:a008698
Walczak H, Degli-Esposti MA, Johnson RS, Smolak PJ, Waugh JY, Boiani N, Timour MS, Gerhart MJ, Schooley KA, Smith CA et al (1997) TRAIL-R2: a novel apoptosis-mediating receptor for TRAIL. EMBO J 16:5386–5397
Walczak H, Krammer PH (2000) The CD95 (APO-1/Fas) and the TRAIL (APO-2 L) apoptosis systems. Exp Cell Res 256:58–66
Wang L, Du F, Wang X (2008) TNF-alpha induces two distinct caspase-8 activation pathways. Cell 133:693–703
Weinlich R, Dillon CP, Green DR (2011) Ripped to death. Trends Cell Biol 21:630–637
Welz PS, Wullaert A, Vlantis K, Kondylis V, Fernandez-Majada V, Ermolaeva M, Kirsch P, Sterner-Kock A, van Loo G, Pasparakis M (2011) FADD prevents RIP3-mediated epithelial cell necrosis and chronic intestinal inflammation. Nature 477:330–334
Wenzel DM, Lissounov A, Brzovic PS, Klevit RE (2011) UBCH7 reactivity profile reveals parkin and HHARI to be RING/HECT hybrids. Nature 474:105–108
Wertz IE, Dixit VM (2010) Regulation of death receptor signaling by the ubiquitin system. Cell Death Differ 17:14–24
Wertz IE, O’Rourke KM, Zhou H, Eby M, Aravind L, Seshagiri S, Wu P, Wiesmann C, Baker R, Boone DL et al (2004) De-ubiquitination and ubiquitin ligase domains of A20 downregulate NF-kappaB signalling. Nature 430:694–699
Wright A, Reiley WW, Chang M, Jin W, Lee AJ, Zhang M, Sun SC (2007) Regulation of early wave of germ cell apoptosis and spermatogenesis by deubiquitinating enzyme CYLD. Dev Cell 13:705–716
Wu GS, Burns TF, McDonald ER 3rd, Jiang W, Meng R, Krantz ID, Kao G, Gan DD, Zhou JY, Muschel R et al (1997) KILLER/DR5 is a DNA damage-inducible p53-regulated death receptor gene. Nat Genet 17:141–143
Wu J, Huang Z, Ren J, Zhang Z, He P, Li Y, Ma J, Chen W, Zhang Y, Zhou X et al (2013) Mlkl knockout mice demonstrate the indispensable role of Mlkl in necroptosis. Cell Res 23:994–1006
Yin Q, Lamothe B, Darnay BG, Wu H (2009) Structural basis for the lack of E2 interaction in the RING domain of TRAF2. Biochemistry 48:10558–10567
Zhang DW, Shao J, Lin J, Zhang N, Lu BJ, Lin SC, Dong MQ, Han J (2009) RIP3, an energy metabolism regulator that switches TNF-induced cell death from apoptosis to necrosis. Science 325:332–336
Zhang H, Zhou X, McQuade T, Li J, Chan FK, Zhang J (2011) Functional complementation between FADD and RIP1 in embryos and lymphocytes. Nature 471:373–376
Zhang L, Blackwell K, Shi Z, Habelhah H (2010) The RING domain of TRAF2 plays an essential role in the inhibition of TNFalpha-induced cell death but not in the activation of NF-kappaB. J Mol Biol 396(3):528–539
Zhao J, Jitkaew S, Cai Z, Choksi S, Li Q, Luo J, Liu ZG (2012) Mixed lineage kinase domain-like is a key receptor interacting protein 3 downstream component of TNF-induced necrosis. Proc Natl Acad Sci U S A 109:5322–5327
Zheng C, Kabaleeswaran V, Wang Y, Cheng G, Wu H (2010) Crystal structures of the TRAF2: cIAP2 and the TRAF1: TRAF2: cIAP2 complexes: affinity, specificity, and regulation. Mol Cell 38:101–113
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer Science+Business Media New York
About this chapter
Cite this chapter
Darding, M., Walczak, H. (2014). Regulation of Death Receptor-Induced Necroptosis by Ubiquitination. In: Shen, HM., Vandenabeele, P. (eds) Necrotic Cell Death. Cell Death in Biology and Diseases. Humana Press, New York, NY. https://doi.org/10.1007/978-1-4614-8220-8_5
Download citation
DOI: https://doi.org/10.1007/978-1-4614-8220-8_5
Published:
Publisher Name: Humana Press, New York, NY
Print ISBN: 978-1-4614-8219-2
Online ISBN: 978-1-4614-8220-8
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)