Abstract
Molecular cytogenetics has played an important role in the elucidation of chromosomal structure, behavior, and evolution in the genus Oryza. Extensive and elaborate analyses on rice chromosomes over the past few decades have uncovered cytological features such as gene locations, genome organization at centromeres and subtelomeres, repeat distributions, and epigenetic modifications. In fact, rice can be considered a model species for chromosome studies due to the available tools and information. Fluorescence in situ hybridization (FISH) on wild species and genomic in situ hybridization (GISH) on interspecific crosses revealed conservation and diversity of genomes within the genus Oryza, providing a better understanding of chromosomal evolution in the genus. Combined with genomics, we expect molecular cytogenetics to continue to be an essential tool to facilitate fundamental knowledge on chromosomes and their evolution and to contribute to breeding strategies.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Long Terminal Repeat
- Bacterial Artificial Chromosome Clone
- Molecular Cytogenetic
- Pachytene Chromosome
- Oryza Species
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The genus Oryza is divided into 24 species based on morphological, cytological, biochemical, and molecular evidence including two cultivated species, O. sativa, Asian cultivated rice, and O. glaberrima, African cultivated rice, and 22 wild species represented by 10 genome types [1] (Table 6.1). Rice (O. sativa) is one of the most important staple crops in the world, and thus a number of cytogenetic studies have been done in rice. In recent decades, molecular cytogenetics, which combines molecular techniques and cytogenetics, has contributed to our understanding of chromosome and genome structure, phylogeny, and genome evolution in the Oryza species. Fluorescence in situ hybridization (FISH) has been an essential tool in molecular cytogenetics to visualize unique and repetitive DNA sequences on chromosomes using epifluorescence microscopy. FISH uses labeled nucleotides incorporated into DNA sequences, called a probe, to hybridize to complementary chromosomal DNA sequences. The probe is detected as a signal by a fluorescent microscope. FISH targets include interphase nuclei, somatic chromosomes, meiotic chromosomes, and extended DNA fibers. Recent advances in FISH allow one to distinguish and identify chromosomes, integrate genetic maps with specific chromosomes, compare physical and genetic distances, analyze distribution of repetitive DNA throughout a genome, associate genes or specific DNA sequences to chromosomal positions on chromosomes (Fig. 6.1), and determine genome origins in hybrids.
Use of FISH to detect ribosomal DNA on mitotic prometaphase chromosomes of O. sativa L. ssp. japonica cv. Nipponbare. Chromosomes were counterstained with DAPI (blue). (a) 45S rDNA is detected on chromosome 9 as green signals. (b) 5S rDNA and BAC B1109A06 containing CentO repeat are detected on chromosome 11 as green and red signals, respectively. Bars represent 5 μm
2 Identification and Cytological Analysis of Individual Chromosomes of Rice
Distinguishing individual chromosomes of rice is very difficult due to their small size, similar morphology, and the lack of reliable banding patterns [2]. However, identification of rice chromosomes was achieved by quantitative analysis of uneven condensation patterns (CP) that appear on mitotic prometaphase chromosomes. Based on CPs and centromeric positions, 332 (92.2 %) out of 360 chromosomes were automatically identified, indicating that CP is a reliable and useful approach for chromosome identification [3]. A quantitative chromosome map of 12 rice chromosomes was developed using a chromosome image analyzing system (CHIAS), showing condensation patterns, total chromosomal length, and arm ratio [4, 5].
FISH using chromosome-specific bacterial artificial chromosome (BAC) clones is also a powerful tool to identify chromosomes. Cheng et al. [6] used 24 chromosomal arm-specific BAC clones as probes for FISH to identify chromosomes at the meiotic pachytene stage. Pachytene chromosomes have higher resolution and more distinct cytological characteristics such as heterochromatin, euchromatin, and centromeres. The heterochromatin distribution pattern is highly conserved between japonica and indica subspecies, and an ideogram of pachytene chromosomes with the distribution of heterochromatin was developed based on the staining pattern of 4′,6-diamidino-2-phenylindole (DAPI) in japonica subspecies [6].
3 Physical Mapping of Low- or Single-Copy Sequences on Rice Chromosomes
Physical mapping of low- or single-copy sequences using FISH is an effective way to know the precise location of the sequences on chromosomes and their relation to chromosomal cytological features. In plants, the minimum length of detectable probes is around 1–3 kb [7–9]. However, the use of small probes has issues with low frequency of detection (e.g., distinguishing hybridization signals from background fluorescence noise) as well as inconsistency of the results [10]. To overcome this obstacle, large-insert genomic clones containing small target DNA sequences have been used successfully to map low- or single-copy DNA sequences in many plant genomes. For example, in rice, several BAC clones closely linked to a specific gene such as Pi-b, Xa-21, and Pi-ta [2] were successfully mapped onto mitotic chromosomes using FISH [7, 11, 12].
The integration of genetic and cytological maps is essential to understand chromosome structure and recombination frequency along chromosomes and to determine the precise location of target DNA sequences. Genetic maps do not reflect the actual physical distance across chromosomes because recombination is a nonrandom process across each chromosome. FISH mapping using large DNA clones containing genetic markers is useful to integrate cytogenetic and genetic maps. The rice genetic and cytogenetic maps have been integrated via 24 chromosomal arm-specific BAC clones containing RFLP markers [6]. Eighteen BAC/PAC clones containing RFLP markers were physically mapped onto rice pachytene chromosomes 10 and 5, respectively, to show the precise localization of each BAC clone on pachytene chromosomes. The comparison between genetic and FISH-based cytological maps illustrated the uneven distribution of genetic recombination along entire chromosomes [13, 14].
4 Repetitive Sequences Maintaining Chromosomal Structure and Function in the Genus Oryza
Plant genomes are composed of a significant fraction of repetitive sequences including tandem and dispersed repeats. Some of these repeats are responsible for maintaining chromosomal structure and function such as centromeres, telomeres, and other heterochromatic regions. Transposable elements—DNA transposons and RNA transposons (i.e., retrotransposons)—exist in all species of the genus Oryza. Among these elements, long terminal repeat (LTR) retrotransposons are the most abundant. LTR retrotransposons can increase the genome size of an organism in a relatively short time period because of their replicative mode of transposition and large element size. It is known that recent amplifications of LTR retrotransposons have contributed to the genome expansion of both O. australiensis (EE) and O. granulata (GG) resulting in genome sizes more than twice the size as all the AA genome species, including O. sativa [15]. The abundance and distribution patterns of LTR retrotransposons differ among families; some retrotransposons are dispersed genome wide, whereas others are concentrated in heterochromatic regions, such as centromeres, pericentromeres, or telomeres [16].
Centromeres are critical sites for sister chromatid cohesion and kinetochore assembly. Centromeres become visible as primary constrictions on mitotic and meiotic chromosomes. Centromeres in plant genomes are often organized into megabase (Mb)-sized blocks, consisting mostly of repetitive sequences, such as satellite tandem repeats and retrotransposons. Because of the density of repetitive sequences, centromere organization is still poorly understood even in sequenced higher organisms. Rice is one of the few species where centromere organization is understood due to the use of cytogenetics to assist in the sequencing of several rice centromeres.
Rice centromeres primarily consist of two types of repetitive sequences, centromeric retrotransposons in rice (CRR), and centromere-specific tandem repeats (CentO [17–19]). CRR elements belong to the Ty3-gypsy superfamily, and FISH confirmed that they are enriched in centromeric regions. Unlike most plant LTR retrotransposons, CRRs are highly conserved in the genus Oryza and across the Poaceae family [17]. Southern blot results indicated that CRRs were present in almost all species of the genus Oryza, including the most distant relative of rice, O. granulata [20]. One exception is O. brachyantha (FF) where CRR elements were absent and their role has likely been replaced by another LTR retrotransposon named FRetro3 [20].
In contrast to centromeric retrotransposons, centromere-specific tandem repeats are variable within and between species of the genus Oryza. CentO is present in the AA, BB, CC, BBCC, CCDD, and EE genomes but not in the FF genome [19, 21, 22]. In O. officinalis (CC), the amount of CentO is quite low and another satellite repeat, CentO-C, is the dominant tandem repeat. FISH results showed that CentO exists at centromeres only in one pair of chromosomes in O. officinalis (CC), and the other ten centromeres contain CentO-C [23]. In O. brachyantha (FF), CentO has been completely replaced by CentO-F at all 12 centromeres and has no sequence similarity with CentO [21].
The size and intensity of FISH signals of repetitive sequences are correlated with the relative abundance of the repetitive sequences at their positions. Based on FISH signals of CentO in rice pachytene chromosomes and fiber-FISH results, the amount of CentO in the 12 chromosomes was estimated to range from 60 kb to 2 Mb [19] in size. Centromere 8 is the smallest and was the first completely sequenced centromere from any multicellular eukaryote [24, 25]. The amount of CentO dramatically changes from species to species, even in orthologous chromosomes (Fig. 6.2). A question arises as to why the amount of CentO is so different between orthologous Oryza chromosomes. Recently, 87 kb of CentO sequence in centromere 8 from the Kasalath variety (indica subspecies) was fully sequenced, and the result suggested that CentO was likely amplified in Kasalath after the divergence of subspecies japonica and indica [26]. However, the difference in size of CentO repeats can be more than fivefold between orthologous centromeres of japonica and indica. The simple sequence amplification model may not fully explain such a large difference after the divergence of these two subspecies from their common ancestor. Deletions of the CentO sequence in one species may have accompanied expansion in the other.
Telomeres serve as physical caps to protect the ends of chromosomes from degradation and fusion with each other. So far, telomere sequences from more than 120 eukaryotes have been identified and are included in the Telomere Database [27]. The telomeric repeat, TTAGGG, is highly conserved in vertebrates, invertebrates, plants, and some fungi. The TTTAGGG motif is abundant in telomeric regions of rice and other species of the genus Oryza [28]. The amount of telomeric repeats is variable among different chromosomes. Mizuno et al. [28] estimated that rice telomeres ranged in size from 5.1 kb in chromosome 7L to 10.8 kb in chromosome 6L by using a combination of the terminal restriction fragment (TRF) southern blot and fiber-FISH methods. In addition to telomeric repeats, 355-bp TrsA tandem repeats were identified in subtelomeric regions of rice and other Oryza species. Not all subtelomeric regions contain TrsA sequence and the TrsA-block sizes vary among rice varieties [29]. The function and origin of subtelomeric repeats are not clear, but it was proposed that they may act as buffer sequence to block the spread of gene silencing at telomeric positions [30]. Recent FISH mapping study of subtelomeric repeats in Oryza species provided interesting insight into the origins of centromeric and subtelomeric repeats. TrsC repeats, which are exclusive to the CC genome, were detected at multiple subtelomeric regions of O. officinalis (CC); however, the same repeats had multiple subtelomeric and centromeric loci in O. rhizomatis [23] (CC).
5 Applications of Extended DNA Fiber FISH
Extended DNA fiber FISH (fiber or EDF-FISH) is an improvement of FISH mapping in terms of spatial resolution down to a few kilobases and detection sensitivity up to 700 bp [31–33]. EDF-FISH has multiple applications including estimation of physical length and copy numbers of repetitive sequence and physical gaps between adjacent sequences (e.g., Fig. 6.3). EDF-FISH using telomere sequences and TrsA revealed the structure of the ends of chromosomes 6 and 12 of rice including the distance between telomere sequence and TrsA, their total lengths, and copy numbers [28, 34, 35]. EDF-FISH also revealed the intermingled structure between CentO and CRR [17, 19]. In the rice sequencing project, EDF-FISH was successfully used to determine the size of unsequenced gaps for chromosomes 1, 4, and 10 [36–38].
6 GISH in Oryza Species
Genomic in situ hybridization (GISH) uses a total genomic DNA from one species as a probe to hybridize with target chromosomal DNA. The extent of hybridization depends on sequence identity or divergence between probe DNA and target chromosomal DNA. GISH allows one to identify the genomic contribution in interspecific hybrids and polyploids, alien chromosomes, introgressed chromosomes and chromosomal regions, and the cytogenetic relationship among related genomes. Table 6.2 summarizes the reports of GISH experiments in a variety of Oryza species. These studies show successful GISH experiments in polyploid Oryza species, indicating that their genomes are sufficiently diverged to be distinguished using GISH. The extent of GISH hybridization on O. latifolia (CCDD) and O. minuta (BBCC), using O. officinalis (CC) as a probe, indicates that the C and D genomes are more closely related to each other than the B and C genomes [39].
GISH can also be useful for breeding. GISH has been used to reveal the genomic constitution, including introgressed or translocated regions of chromosomes of interspecific hybrids and derived lines [40–44].
7 Detection of Epigenetic Modification on Chromosomes
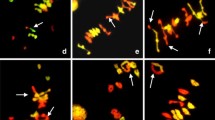
Heterochromatin plays a significant role in the suppression of genes and transposable elements as well as the maintenance of chromosome structure. The epigenetic networks of molecular interactions, including DNA methylation, histone modification, and recruitment of protein complexes, are associated with heterochromatin formation and maintenance. Several studies have shown the association of particular epigenetic modifications with gene silencing and heterochromatin. For example, in Arabidopsis, dimethylated histone H3K9 has been shown to be a critical mark for DNA methylation and gene silencing [45]. Both methylated DNA and dimethylated histone H3K9 are also prominent epigenetic marks for heterochromatic regions, indicating their potential roles in heterochromatin formation and maintenance [46]. Rice has the patterns of epigenetic modifications consistent with Arabidopsis. Methylcytosine immunoprecipitation (mCIP) and immunostaining using an anti-5-methylcytosine antibody revealed that the chromosomal distribution of DNA methylation is coincident with heterochromatin on rice meiotic chromosomes [47]. Immunostaining using anti-dimethylated H3K9 showed that dimethylated H3K9 signals were detected along rice pachytene chromosomes, especially at pericentromeric regions, consistent with the DNA methylation pattern on rice pachytene chromosomes (Fig. 6.4) [48]. Figure 6.4d shows a pachytene chromosome where the signal of dimethylated H3K9 is enriched at condensed regions within each chromomere.
Immunostaining results of histone dimethylated H3K9 [48]. This image is modified from Iwata et al. (a) Meiotic chromosomes at early pachytene stage counterstained with DAPI. (b, d) Signals of dimethylated H3K9. (c) A pachytene chromosome counterstained with DAPI. Bar represents 5 μm
8 Conclusion and Prospectives
Rice has been and continues to be a model system for genetic, genomic, and cytogenetic studies. With the availability of genetic resources spanning the entire genus [49], studies have led to greater knowledge of chromosomal structure and behavior, positions of interesting genes, centromere structure and function, epigenetic modifications, and chromosomal evolution in the genus Oryza. The development of FISH techniques and their applications have clearly broadened our fundamental knowledge of chromosomal biology. The combination of molecular cytogenetics and high-throughput sequencing techniques allows one to explore new facets of chromosome behavior and the effect of chromosome structure and positioning on gene function and patterns of epigenetic modifications, associated with chromosomal structures. Molecular cytogenetics research in the wild relatives of rice, spanning 15–20 MY of evolution, will illuminate the effects of evolutionary pressures on chromosome structure and behavior. We expect that the primary component of plant genomes, retrotransposons, will be a major modeling component of chromosomes, but it remains to be seen either at a chromosomal level or even at a genome level how these genomic “parasites” affect chromosomal and genetic adaptation and evolution.
References
Vaughan DA, Morishima H, Kadowaki K (2003) Diversity in the Oryza genus. Curr Opin Plant Biol 6(2):139–146
Kurata N, Fukui K (2003) Chromosome research in genus Oryza. In: Nanda JS, Sharma SD (eds) Monograph in genus Oryza. Science Publishers, New Hampshire, pp 213–261
Kamisugi Y, Furuya N, Iijima K, Fukui K (1993) Computer-aided automatic identification of rice chromosomes by image parameters. Chromosome Res 1(3):189–196
Fukui K (1996) Advances in rice chromosome research, 1990–95. In: Rice genetics, III: proceedings of the third international rice genetics. IRRI, Manila, Philippines, pp 117–130
Fukui K, Iijima K (1991) Somatic chromosome map of rice by imaging methods. Theor Appl Genet 81(5):589–596
Cheng Z, Buell CR, Wing RA, Gu M, Jiang J (2001) Toward a cytological characterization of the rice genome. Genome Res 11(12):2133–2141
Ohmido N, Akiyama Y, Fukui K (1998) Physical mapping of unique nucleotide sequences on identified rice chromosomes. Plant Mol Biol 38(6):1043–1052
Wang CJR, Harper L, Cande WZ (2006) High-resolution single-copy gene fluorescence in situ hybridization and its use in the construction of a cytogenetic map of maize chromosome 9. Plant Cell 18(3):529–544
Danilova TV, Birchler JA (2008) Integrated cytogenetic map of mitotic metaphase chromosome 9 of maize: resolution, sensitivity, and banding paint development. Chromosoma 117(4):345–356
Jiang JM, Gill BS (2006) Current status and the future of fluorescence in situ hybridization (FISH) in plant genome research. Genome 49(9):1057–1068
Jiang JM, Gill BS, Wang GL, Ronald PC, Ward DC (1995) Metaphase and interphase fluorescence in situ hybridization mapping of the rice genome with bacterial artificial chromosomes. Proc Natl Acad Sci U S A 92(10):4487–4491
Nakamura S, Asakawa S, Ohmido N, Fukui K, Shimizu N, Kawasaki S (1997) Construction of an 800-kb contig in the near-centromeric region of the rice blast resistance gene Pi-ta 2 using a highly representative rice BAC library. Mol Gen Genet 254(6):611–620
Cheng ZK, Presting GG, Buell CR, Wing RA, Jiang JM (2001) High-resolution pachytene chromosome mapping of bacterial artificial chromosomes anchored by genetic markers reveals the centromere location and the distribution of genetic recombination along chromosome 10 of rice. Genetics 157(4):1749–1757
Kao FI, Cheng YY, Chow TY et al (2006) An integrated map of Oryza sativa L. Chromosome 5. Theor Appl Genet 112(5):891–902
Piegu B, Guyot R, Picault N et al (2006) Doubling genome size without polyploidization: dynamics of retrotransposition-driven genomic expansions in Oryza australiensis, a wild relative of rice. Genome Res 16(10):1262–1269
Kumar A, Bennetzen JL (1999) Plant retrotransposons. Annu Rev Genet 33:479–532
Dong F, Miller JT, Jackson SA, Wang GL, Ronald PC, Jiang J (1998) Rice (Oryza sativa) centromeric regions consist of complex DNA. Proc Natl Acad Sci U S A 95(14):8135–8140
Miller JT, Dong F, Jackson SA, Song J, Jiang J (1998) Retrotransposon-related DNA sequences in the centromeres of grass chromosomes. Genetics 150(4):1615–1623
Cheng Z, Dong F, Langdon T et al (2002) Functional rice centromeres are marked by a satellite repeat and a centromere-specific retrotransposon. Plant Cell 14(8):1691–1704
Gao D, Gill N, Kim HR et al (2009) A lineage-specific centromere retrotransposon in Oryza brachyantha. Plant J 60(5):820–831
Lee HR, Zhang W, Langdon T et al (2005) Chromatin immunoprecipitation cloning reveals rapid evolutionary patterns of centromeric DNA in Oryza species. Proc Natl Acad Sci U S A 102(33):11793–11798
Zhang WL, Yi CD, Bao WD et al (2005) The transcribed 165-bp CentO satellite is the major functional centromeric element in the wild rice species Oryza punctata. Plant Physiol 139(1):306–315
Bao W, Zhang W, Yang Q et al (2006) Diversity of centromeric repeats in two closely related wild rice species, Oryza officinalis and Oryza rhizomatis. Mol Genet Genomics 275(5):421–430
Nagaki K, Cheng Z, Ouyang S et al (2004) Sequencing of a rice centromere uncovers active genes. Nat Genet 36(2):138–145
Wu J, Yamagata H, Hayashi-Tsugane M et al (2004) Composition and structure of the centromeric region of rice chromosome 8. Plant Cell 16(4):967–976
Wu J, Fujisawa M, Tian Z et al (2009) Comparative analysis of complete orthologous centromeres from two subspecies of rice reveals rapid variation of centromere organization and structure. Plant J 60(5):805–819
Podlevsky JD, Bley CJ, Omana RV, Qi X, Chen J (2007) The telomerase database. Nucleic Acids Res 36:D339–D343
Mizuno H, Wu J, Kanamori H et al (2006) Sequencing and characterization of telomere and subtelomere regions on rice chromosomes 1S, 2S, 2L, 6L, 7S, 7L and 8S. Plant J 46(2):206–217
Ohmido N, Kijima K, Akiyama Y, de Jong JH, Fukui K (2000) Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Mol Gen Genet 263(3):388–394
Mizuno H, Wu J, Katayose Y, Kanamori H, Sasaki T, Matsumoto T (2008) Characterization of chromosome ends on the basis of the structure of TrsA subtelomeric repeats in rice (Oryza sativa L.). Mol Genet Genomics 280(1):19–24
Fransz PF, Alonso-Blanco C, Liharska TB, Peeters AJ, Zabel P, de Jong JH (1996) High-resolution physical mapping in Arabidopsis thaliana and tomato by fluorescence in situ hybridization to extended DNA fibres. Plant J 9(3):421–430
Jackson SA, Wang ML, Goodman HM, Jiang J (1998) Application of fiber-FISH in physical mapping of Arabidopsis thaliana. Genome 41(4):566–572
Cheng Z, Buell CR, Wing RA, Jiang J (2002) Resolution of fluorescence in-situ hybridization mapping on rice mitotic prometaphase chromosomes, meiotic pachytene chromosomes and extended DNA fibers. Chromosome Res 10(5):379–387
Fukui K, Ohmido N, Kijima K, Akiyama Y, de Jong JH (2000) Quantification of total genomic DNA and selected repetitive sequences reveals concurrent changes in different DNA families in indica and japonica rice. Mol Gen Genet 263(3):388–394
Ohmido N, Kijima K, Ashikawa I, de Jong JH, Fukui K (2001) Visualization of the terminal structure of rice chromosomes 6 and 12 with multicolor FISH to chromosomes and extended DNA fibers. Plant Mol Biol 47(3):413–421
Feng Q, Zhang YJ, Hao P et al (2002) Sequence and analysis of rice chromosome 4. Nature 420(6913):316–320
Sasaki T, Matsumoto T, Yamamoto K et al (2002) The genome sequence and structure of rice chromosome 1. Nature 420(6913):312–316
Rice Chromosome 10 Sequencing Consortium (2003) In-depth view of structure, activity, and evolution of rice chromosome 10. Science 300(5625):1566–1569
Fukui K, Shishido R, Kinoshita T (1997) Identification of the rice D-genome chromosomes by genomic in situ hybridisation. Theor Appl Genet 95(8):1239–1245
Shishido R, Ohmido N, Fukui K (2001) Chromosome painting as a tool for rice genetics and breeding. Methods Cell Sci 23(1–3):125–132
Yan H, Liu G, Cheng Z, Min S, Zhu L (2001) Characterization of euploid backcross progenies derived from interspecific hybrids between Oryza sativa and O. eichingeri by restriction fragment length polymorphism (RFLP) analysis and genomic in situ hybridization (GISH). Genome 44(1):86–95
Tan G, Jin H, Li G, He R, Zhu L, He G (2005) Production and characterization of a complete set of individual chromosome additions from Oryza officinalis to Oryza sativa using RFLP and GISH analyses. Theor Appl Genet 111(8):1585–1595
Jin H, Tan G, Brar DS et al (2006) Molecular and cytogenetic characterization of an Oryza officinalis-O. sativa chromosome 4 addition line and its progenies. Plant Mol Biol 62(4–5):769–777
Xiong ZY, He GC, Tan GX, He GY, Song YC (2006) Cytogenetic comparisons between A and G genomes in Oryza using genomic in situ hybridization. Cell Res 16(3):260–266
Jackson JP, Johnson L, Jasencakova Z et al (2004) Dimethylation of histone H3 lysine 9 is a critical mark for DNA methylation and gene silencing in Arabidopsis thaliana. Chromosoma 112(6):308–315
Jasencakova Z, Soppe WJJ, Meister A, Gernand D, Turner BM, Schubert I (2003) Histone modifications in Arabidopsis: high methylation of H3 lysine 9 is dispensable for constitutive heterochromatin. Plant J 33(3):471–480
Yan HH, Kikuchi S, Neumann P et al (2010) Genome-wide mapping of cytosine methylation revealed dynamic DNA methylation patterns associated with genes and centromeres in rice. Plant J 63(3):353–365
Iwata A (2008) Cytological research of rice chromosomes. Kobe University Graduation thesis, pp 1–51
Wing RA, Ammiraju JS, Luo M et al (2005) The Oryza Map Alignment Project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol Biol 59(1):53–62
International Rice Sequencing Project (2005) The map-based sequence of the rice genome. Nature 436(7052):793–800
Ammiraju JS, Luo M, Goicoechea JL et al (2006) The Oryza bacterial artificial chromosome library resource: construction and analysis of 12 deep-coverage large-insert BAC libraries that represent the 10 genome types of the genus Oryza. Genome Res 16(1):140–147
Iyengar GAS, Sen SK (1978) Nuclear-DNA content of several wild and cultivated Oryza species. Environ Exp Bot 18(4):219–224
Uozu S, Ikehashi H, Ohmido N, Ohtsubo H, Ohtsubo E, Fukui K (1997) Repetitive sequences: cause for variation in genome size and chromosome morphology in the genus Oryza. Plant Mol Biol 35(6):791–799
Martínez CP, Arumuganathan K, Kikuchi H, Earle ED (1994) Nuclear DNA content of ten rice species as determined by flow cytometry. Jpn J Genet 69:513–523
Acknowledgements
We would like to acknowledge and thank the USA National Science Foundation for funding to support this work (NSF DBI 105048).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Iwata, A., Gao, D., Ohmido, N., Jackson, S.A. (2013). Molecular Cytogenetics of Rice and Its Wild Relatives. In: Zhang, Q., Wing, R. (eds) Genetics and Genomics of Rice. Plant Genetics and Genomics: Crops and Models, vol 5. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-7903-1_6
Download citation
DOI: https://doi.org/10.1007/978-1-4614-7903-1_6
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-7902-4
Online ISBN: 978-1-4614-7903-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)