Abstract
PR, IN, and RT are the only enzymes encoded by the HIV genome, and all are targeted by antiviral therapeutic agents (De Clercq 2009). HIV PR cleaves viral polyprotein precursors to produce mature virus and is targeted by PR inhibitors saquinavir, ritonavir, and several others. IN is required for assimilation of viral DNA into the genome of the infected cell and is targeted by raltegravir. A second IN inhibitor, elvitegravir, is currently in the latter stages of clinical trials. RT possesses two enzymatic activities: DNA polymerase and RNase H. The former is targeted by both NRTIs (e.g., didanosine, or 2′,3′-dideoxyinosine) and NNRTIs (e.g., nevirapine, efavirenz) (Ruane and DeJesus 2004). Of the 25 anti-HIV compounds in clinical use today, 12 target the multifunctional RT enzyme. However, none of these inhibitors target the RNase H activity of RT, despite it being absolutely essential for synthesis of preintegrative viral DNA (Schatz et al. 1989). In this chapter, various aspects of HIV-1 RT-associated RNase H and its inhibition will be discussed, including (i) the structure of HIV-1 RT, with emphasis on the RNase H domain; (ii) the two-metal-ion-dependent mechanism of RNase H cleavage; and (iii) efforts and progress toward developing potent and specific inhibitors of the lone untargeted HIV enzymatic function.
Conflict of interest statement: None declared
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
PR, IN, and RT are the only enzymes encoded by the HIV genome, and all are targeted by antiviral therapeutic agents (De Clercq 2009). HIV PR cleaves viral polyprotein precursors to produce mature virus and is targeted by PR inhibitors saquinavir, ritonavir, and several others. IN is required for assimilation of viral DNA into the genome of the infected cell and is targeted by raltegravir. A second IN inhibitor, elvitegravir, is currently in the latter stages of clinical trials. RT possesses two enzymatic activities: DNA polymerase and RNase H. The former is targeted by both NRTIs (e.g., didanosine, or 2′,3′-dideoxyinosine) and NNRTIs (e.g., nevirapine, efavirenz) (Ruane and DeJesus 2004). Of the 25 anti-HIV compounds in clinical use today, 12 target the multifunctional RT enzyme. However, none of these inhibitors target the RNase H activity of RT, despite it being absolutely essential for synthesis of preintegrative viral DNA (Schatz et al. 1989). In this chapter, various aspects of HIV-1 RT-associated RNase H and its inhibition will be discussed, including (i) the structure of HIV-1 RT, with emphasis on the RNase H domain; (ii) the two-metal-ion-dependent mechanism of RNase H cleavage; and (iii) efforts and progress toward developing potent and specific inhibitors of the lone untargeted HIV enzymatic function.
2 Structure of HIV-1 RT and the RNase H Domain
HIV-1 RT is a heterodimer of 66- and 51-kD subunits (p66 and p51, respectively) cleaved from the gag-pol precursor by HIV-1 PR during virus maturation (Di Marzo Veronese et al. 1986). Many X-ray crystal structures of HIV-1 RT have been resolved, including apoenzyme and enzyme liganded with double-stranded nucleic acid, dNTP, and/or any of several RT inhibitors (Huang et al. 1998; Jacobo-Molina et al. 1993; Kohlstaedt et al. 1992; Ren et al. 2001; Rodgers et al. 1995; Sarafianos et al. 2001). Although amino acids 1–440 are common to p66 and p51, the two subunits assume quite different tertiary structures in the context of heterodimeric RT (Fig. 7.1). The larger subunit houses the DNA polymerase and RNase H active centers of the enzyme toward its amino- and carboxy-termini, respectively (Rothwell and Waksman 2005). The polymerase domain resembles a grasping right hand, comprised of fingers (residues 1–85, 118–155), palm (86–117, 155–237), and thumb (238–318) subdomains, and is linked to the RNase H domain via a connection subdomain (residues 319–426) (Fig. 7.1a). In contrast, p51 is relatively compact, serving primarily to support the more extended structure of its catalytic counterpart (Fig. 7.1b). Together, the two subunits form a template-primer binding cleft capable of accommodating dsDNA, dsRNA, or an RNA/DNA hybrid, all of which are required for synthesis of the preintegrative DNA intermediate.
Structure of HIV-1 RT in complex with an RNA/DNA hybrid (Sarafianos et al. 2001). RNA and DNA strand ladders are depicted in navy and blue gray, respectively. RT subdomains in (a) p66 and (b) p51 are highlighted
The RNase H domain (p66 residues 427–560) is comprised of a central five-stranded mixed β-sheet surrounded by four α-helices and eight connecting loops (Davies et al. 1991). It is stabilized by interactions with the p66 connection subdomain, as well as with the p51 thumb and connection subdomains (Kohlstaedt et al. 1992), rendering the HIV-1 RNase H domain less flexible than its counterparts in Mo-MLVRT (Das and Georgiadis 2004). The RNase H active site is located on a highly solvent-exposed surface facing the same direction as the interior of the template-primer binding cleft. It is comprised of four highly conserved acidic residues (D443, E478, D498, and D549) that bind the Mg ions necessary for catalysis, as well as a flexible “His loop” near the interface with the p51 thumb. This DEDD motif is common to members of the polynucleotidyltransferase family of enzymes, which includes relatives such as E. coli and B. halodurans RNases H, HIV IN, E. coli RuvC, and the PIWI domain of argonaute, the catalytic component of the RISC complex (Ma et al. 2005; Song et al. 2004).
In X-ray crystal structures in which RT is in complex with duplex DNA (Huang et al. 1998; Jacobo-Molina et al. 1993), RT-nucleic acid contacts occur at multiple sites throughout the polymerase domain, especially around the polymerase active center, and involving the so-called primer and template grip residues. As with most nonspecific nucleic acid-binding proteins, contacts outside of the polymerase active center occur primarily along the sugar-phosphate backbones of both strands of nucleic acid. Duplex DNA bound in the polymerase domain is constrained in an A-form geometry; however, it relaxes into a B-form conformation as it extends past the connection subdomain and RNase H domain. The transition from A- to B-form is marked by a ~40° bend in the substrate and occurs 6–8 bp from the primer 3′ terminus. The polymerase and RNase H domains are separated by approximately 18 bp of dsDNA – a structural observation confirmed by biochemical experimentation (Gotte et al. 1998).
Crystal structures of RT in complex with dsDNA and an incoming dNTP (Huang et al. 1998) or an RT inhibitor (Das et al. 2012; Kohlstaedt et al. 1992; Ren et al. 2001) have greatly clarified how these ligands/inhibitors are bound by the enzyme. Binding sites for dNTPs and NRTIs have been shown to involve primarily fingers and palm subdomain residues. In contrast, the allosteric NNRTIs bind at a site hidden at the base of the p66 thumb. This site is not apparent in the absence of inhibitor but is revealed only when the thumb is displaced by NNRTI binding (Das et al. 2012; Kohlstaedt et al. 1992). Interestingly, the NNRTI binding site does not exist at all in HIV-2 RT, rendering this class of inhibitors ineffective against HIV-2 (Shih et al. 1991). While primarily targeting the polymerase activity of RT, NNRTIs have also been shown to affect RNase H cleavage via an allosteric mechanism, an inhibitory activity that may contribute to antiviral function of this class of inhibitors (Hang et al. 2007).
To date, only a single X-ray crystal structure has been solved for RT in complex with an RNA/DNA hybrid, which in this case contains the HIV-1 3′ PPT together with flanking regions of the viral genomic sequence (Sarafianos et al. 2001). While this segment of RNA is alternately recognized as both template and primer during the course of viral DNA synthesis, it is oriented in the position of template in the co-crystal structure. Several contacts between RT and RNA 2′ hydroxyl groups are revealed in this structure, as are contacts between the DNA primer strand and RT residues in the vicinity of the connection and RNase H domains. These contacts are made by residues collectively referred to as the RNase H primer grip, including G359, A360, H361, T473, N474, Q475, K476, Y501, and I505 from the p66 subunit as well as K395 and E396 of p51. Although the apparent role of the RNase H primer grip is to guide the hybrid toward the RNase H active center, the template is not properly aligned for hydrolysis in this RT co-crystal structure. Specifically, as is the case with DNA template strands in RT-dsDNA co-crystal structures, the RNA strand in the RT-RNA/DNA structure does not pass into the RNase H catalytic center, and the purported scissile phosphate resides ~4 Å from the optimal location for cleavage. This may be because the hybrid duplex assumes a DNA-like B-form conformation in the vicinity of the connection subdomain and RNase H domain, contains localized disruptions in rA:dT base pairing, and is unresolved (and base pairing is likely disrupted altogether) as it emerges past the RNase H domain.
Structures of the isolated HIV-1 RNase H domain, as well as E. coli and B. halodurans RNases H, have also been resolved (Davies et al. 1991; Katayanagi et al. 1990; Nowotny et al. 2005). Unlike its E. coli counterpart, the HIV-1 RNase H domain lacks a basic loop-helix motif (α-helix C) that in the bacterial and some retroviral enzymes contributes to substrate binding. The HIV element relies instead upon extensive contacts in the polymerase domain in addition to those within the RNase H primer grip. Consequently, the isolated HIV-1 domain is highly deficient in substrate binding and is only weakly active or inactive as an RNase H (Cirino et al. 1993; Hansen et al. 1988). The addition of flanking connection subdomain residues or a polyhistidine tag, insertion of an E. coli RNase H α-helix C, and/or use of Mn+2 instead of Mg+2 in RNase H assays restore activity to the isolated HIV-1 domain, allowing it to be used for RNase H inhibitor screening (Hansen et al. 1988; Keck and Marqusee 1995; Stahl et al. 1994).
Finally, active site mutants of B. halodurans RNase H were crystallized in complex with an RNA/DNA hybrid, greatly elucidating the mechanisms of substrate binding and RNase H cleavage (Nowotny et al. 2005). In contrast to substrate within the HIV-1 RT-RNA/DNA co-crystal structure, the B. halodurans RNase H substrate largely assumes an A-like or mixed conformation. Moreover, a four-residue motif designated the phosphate-binding pocket (PBP) was shown in the bacterial enzyme to bind and distort the DNA strand two base pairs from the scissile phosphate, thereby restricting substrates to RNA/DNA hybrids alone. Homology between the B. halodurans and HIV-1 PBPs is incomplete, however, with only T473 and Q475 likely to serve a similar role as the HIV-1 enzyme. This may explain why the cleavage specificity of the HIV-1 RNase H is somewhat relaxed, with RT capable of cleaving one strand in an RNA duplex in certain specialized substrates (Gotte et al. 1995).
3 Mechanism of RNase H Cleavage
During the course of viral DNA synthesis, HIV-1 RT encounters numerous hybrid nucleic acid duplexes in many different structural contexts (Telesnitsky and Goff 1997). RNA hydrolysis occurs both throughout the course of minus-strand DNA synthesis and during more specialized events such as minus-strand and plus-strand transfer, PPT selection, and minus- and plus-strand primer removal. Detailed reviews encompassing these events, structural variations among RNA/DNA hybrids, and the manner in which RT accommodates these substrates are available elsewhere (Champoux and Schultz 2009; Rausch and Le Grice 2004; Schultz and Champoux 2008). Here, however, a brief summary of the different modes of RNase H cleavage is provided.
“Polymerase-dependent” cleavage refers to hydrolytic events catalyzed during the course of DNA synthesis by the polymerizing enzyme (Furfine and Reardon 1991; Peliska and Benkovic 1992). These generally occur ~18 bp from the primer terminus either prior to initiation, during pausing, or after termination of synthesis while the enzyme remains bound to the nascent hybrid. Hydrolysis products generated by this mode of cleavage are both consistent with and identified by the spacing between polymerase and RNase H active sites in HIV-1 RT (~18 bp). This spacing suggests that polymerase-dependent cleavage occurs while the polymerase domain is engaged at or near the 3′ terminus of the DNA primer. Moreover, the tendency for most hydrolytic events to occur when RT is not actively synthesizing DNA suggests that nucleotide incorporation/enzyme translocation and RNase H cleavage do not occur concurrently. The notion that HIV-1 RT cannot simultaneously catalyze both DNA synthesis and RNase H cleavage is consistent with a structural model of RT in complex with an RNA/DNA hybrid spanning the two active sites (Nowotny et al. 2007).
In arrested RT-RNA/DNA complexes, “polymerase-dependent” cleavage events can be subdivided into those occurring “pre-” and “post-translocation,” where translocation refers to the incremental advance of the enzyme that occurs upon incorporation of a dNTP into the primer strand (Gotte et al. 2010). More specifically, incorporation of (i) a ddNTP at the 3′ terminus of the primer strand followed by (ii) inclusion of the next dNTP to be incorporated in the reaction mixture locks the RT-RNA/DNA complex into the “post-translocation” state. Conversely, inclusion of the pyrophosphate analog phosphonoformic acid (PFA, foscarnet) in the reaction mixture stabilizes the “pre-translocation” state of the enzyme-substrate complex. In both “pre-” and “post-translocation” complexes, RNase H-mediated hydrolysis occurs ~18 bp from the primer terminus; however, the precise cleavage site in the former complex is one nucleotide further removed from the primer terminus than in the latter. This observation becomes particularly useful when examining the mechanisms of RNase H inhibition and/or the functional relationship(s) between RNase H and DNA polymerase inhibitors.
Likewise, in the absence of dNTPs, ddNTPs, or other modulators of polymerase domain function, cleavage of the RNA strand occurs primarily ~18 bp from the recessed 3′ terminus, again suggesting engagement of the primer terminus by the polymerase domain of the enzyme (Champoux and Schultz 2009; Furfine and Reardon 1991). This “primary” cleavage is followed by one or more “secondary” hydrolytic events 7–12 bp from the primer terminus (Palaniappan et al. 1996). The latter are also classified as “polymerase-independent” cleavage events, since the spacing between the primer terminus and site(s) of secondary cleavage is not consistent with their simultaneous occupancy within the two active sites of the enzyme (Furfine and Reardon 1991; Peliska and Benkovic 1992).
“Primary” cleavage events are also referred to as “3′-end-directed” cleavages since they occur when the polymerase domain is oriented over the recessed 3′ terminus of the primer strand. Conversely, when the substrate contains a 3′ DNA overhang, cleavage is observed 13–19 nt from the recessed RNA 5′ terminus (DeStefano et al. 1993; Palaniappan et al. 1996). These hydrolytic events are dubbed “5′-end-directed” cleavages since the polymerase domain is located over the DNA strand opposite the recessed RNA 5′ terminus. Moreover, on longer substrates, “internal” cleavage occurs at sites well removed from the 3′ and 5′ termini of either strand (Schultz et al. 2004). While less efficient than either end-directed mode of cleavage, “internal” cleavage is likely at least partly responsible for clearance of genomic RNA following minus-strand DNA synthesis. For this mode of substrate binding and hydrolysis in particular, RNA sequence appears to play an important role in cleavage site selection. While not absolute, a set of specificity rules for predicting the sites at which RNase H-mediated hydrolysis is likely to occur has been established, based on hydrolysis patterns obtained from cleavage of a wide variety of RNA/DNA hybrids (Champoux and Schultz 2009; Schultz and Champoux 2008; Schultz et al. 2004).
Nucleic acid sequence and structure, as well as numerous protein-nucleic acid contacts throughout RT and the template-primer, serve as determinants affecting both substrate binding and RNase H cleavage specificity and efficiency. Nevertheless, the chemistry of catalysis is likely common to all modes of RNase H-mediated hydrolysis. The cleavage mechanism was derived from biochemical experiments (Klumpp et al. 2003) and validated in a series of B. halodurans RNase H-RNA/DNA co-crystal structures in which enzymes containing mutations in conserved active site residues were used to capture various catalytic intermediates (Nowotny et al. 2005). Moreover, a molecular model integrating the catalytic mechanisms of B. halodurans RNase H and human RNase H1 into an HIV RT-RNA/DNA complex has been generated (Nowotny et al. 2007), although the structural coordinates for this model are not publicly available. Here, detailed structures of the HIV-1 RT-associated and B. halodurans RNase H active sites are provided in Fig. 7.2a, b, respectively.
RNase H active sites. (a) Image of RT-associated RNase H active site generated using RT-RNA/DNA co-crystal structure coordinates (Sarafianos et al. 2001). Catalytic residues are highlighted. The scissile phosphate (wheat spheres) of the RNA strand (navy ladder) is not properly positioned for cleavage in this complex nor are the two metal-ion cofactors present. (b) Active site of Bacillus halodurans RNase H (Nowotny et al. 2005). Catalytic amino acids are labeled together with the homologous HIV residues (parentheses), as are the A- and B-site Mg ions. The crystallized enzyme was engineered to contain a D192N mutation in order to ensure that the RNA strand would not be hydrolyzed
Extrapolating the B. halodurans RNase H mechanism to HIV-1 RT-associated RNase H, two Mg+2 ions (A and B) are coordinated in part by conserved acidic residues at adjacent sites in the RNase H active center: Mg+2 A by D443 and D549, and Mg+2 B by D443, E478, and D498. Other coordination partners for these ions at various stages of the reaction include water and both bridging and non-bridging oxygen atoms of the scissile phosphodiester bond. Involvement of the substrate in metal-ion coordination suggests that Mg+2 binding at one or both sites may be substrate dependent, a notion supported by an isothermal calorimetric study of HIV-1 RT demonstrating that only one Mg ion is bound at the RNase H active center in the apoenzyme (Cowan et al. 2000).
In the initial stage of the cleavage reaction, the two catalytic metal ions are 4 Å apart. Mg ion A is coordinated in perfect octahedral geometry and activates a water molecule for nucleophilic attack on the scissile phosphate. In contrast, the coordination geometry of Mg ion B is slightly distorted, perhaps serving to destabilize the enzyme-substrate complex and drive it toward catalysis. Nucleophilic attack on the scissile phosphate ensues, bringing the Mg ions closer together (3.5 Å) and creating a pentavalent phosphate intermediate. This intermediate is quickly converted to the 5′-phosphate and 3′-hydroxyl cleavage products, completing the Sn2 substitution reaction and once again separating the Mg ions (4.8 Å). For B. halodurans RNase H, it was speculated that repositioning of residue D188 facilitated metal-ion and/or product displacement once the cleavage reaction was complete. The equivalent position in HIV-1 RT-associated RNase H is occupied by H539, a residue at the apex of the flexible “His loop” demonstrated by mutational analysis to play an important but nonessential role in catalysis (Schatz et al. 1989).
4 Inhibitors of HIV-1 RT-Associated RNase H
Although there are 12 HIV-1 RT inhibitors in clinical use (De Clercq 2009), with many others available for laboratory applications, development of a potent and specific RNase H inhibitor has proven elusive. In contrast, HIV-1 IN, which possesses a DEDD active center motif homologous to that of RNase H with similar nucleolytic function (Nowotny 2009), has been effectively targeted by antiviral drugs. The difficulty in targeting RNase H, an activity shown to be as essential to virus replication as that of any other viral enzyme (Schatz et al. 1989), may lie in the relatively open topology of its active center compared to its viral enzyme counterparts (Fig. 7.3). While potential inhibitor binding pockets exist in and around the RNase H domain, there are few near the active center, suggesting that effective small molecule inhibition of RNase H activity will require binding directly either to the catalytic metal ions present at the enzyme active site or to a remote site for allosteric inhibition of RNase H.
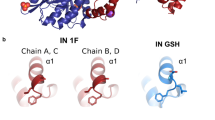
Comparison of RNase H, DNA polymerase, and integrase active sites. (a) Surface representation of the RNase H active site from the RT-RNA/DNA co-crystal structure (Sarafianos et al. 2001). Locations of catalytic and other active site residues are highlighted. Although active site metal ions are not present in the co-crystal, the relatively open structure of the RT-associated RNase H active site is revealed. (b) Polymerase active site of RT in complex with dsDNA and an incoming dNTP (Huang et al. 1998). Select p66 subdomains are highlighted, as are the DNA template (navy ladder), DNA primer (blue-gray, semitransparent ladder), and catalytic Mg ions (spheres). The incoming dNTP (yellow sticks) coordinates the two active site metal ions, is nestled tightly among fingers and palm subdomain residues, and is engaged in a stacking interaction with the 3′-terminal primer nucleotide. Note that NRTIs would be expected to be accommodated in the same, or a similar, manner prior to incorporation into nascent DNA. To facilitate visualization of the polymerase active site, the p66 thumb subdomain has been removed. (c) Surface representation of the prototype spuma viral integrase active center in a complex containing divalent metal ions (blue-white spheres), dsDNA (navy, blue-gray ladders), and the metal-binding inhibitor raltegravir (Hare et al 2010). The inhibitor is shown to coordinate both active site metal ions, form close contacts with integrase residues P214 and Y212, and form π-stacking interactions with the invariant C-A nucleotide tandem at the 3′ terminus of the short DNA strand
4.1 Active Site Inhibitors
Direct targeting of the RNase H active site typically requires that compounds contain a three-oxygen pharmacophore that, in conjunction with the four acidic active site residues, effectively and simultaneously coordinates the two catalytic Mg ions (Klumpp and Mirzadegan 2006). Binding the active site metal ions in this manner would be expected to prevent accommodation of substrate within the RNase H domain, inhibit catalytic chemistry, and/or prevent hydrolysis. Optimal ionic interaction between metal ions and inhibitor requires that compounds have at least one negative charge, preferably on the bridging oxygen that interacts with both ions simultaneously. Unfortunately, compounds with such a charge would also be expected to have limited cellular permeability – a deficiency commonly observed in this class of inhibitors. Moreover, in contrast to other enzymes that utilize tandem metal ions, the relatively open surface of the RNase H domain limits secondary interactions between enzyme and inhibitor that might be expected to increase binding affinity and inhibitor specificity (Fig. 7.3). Despite these challenges, a number of submicromolar inhibitors that directly target the RNase H active site have been developed (Fig. 7.4).
4.1.1 N-Hydroxyimides
Originally identified as inhibitors of influenza virus endonuclease (Parkes et al. 2003), N-hydroxyimides were the first compounds to be specifically selected and/or designed to target the HIV-1 RNase H active center (Klumpp et al. 2003). As is now the standard for this class of inhibitor, N-hydroxyimides contain three oxygen ligands optimally oriented to simultaneously interact with two metal ions spaced ~4 Å apart. In the original experiments, compounds were designed to have minimal molecular weight, complexity, and potential for interaction with protein so that their inhibitory potency would be based solely on their interaction with metal ions. Moreover, it was intended that binding to enzymes requiring fewer than two metal ions should be minimal, and catalytic activity should not be affected. E. coli RNase H was to serve as a control in this regard, as this enzyme is thought to utilize a one-metal-ion-dependent mechanism for catalysis (Keck et al. 1998; Tsunaka et al. 2003, 2005).
As the dual metal-ion-binding model predicted, the prototype compound (1,2-hydroxy-4H-isoquinoline-1,3-dione) potently inhibited HIV RNase H activity (IC50 = 0.6–1 μM) yet was completely ineffective against E. coli RNase H (IC50 > 50 μM) (Hang et al. 2004; Klumpp et al. 2003). This specificity was ascribed to a difference in the number of metal ions present at the active sites of the two enzymes, although it is possible that differences in substrate affinity and turnover between HIV-1 and E. coli RNases H were influential as well. In chemical substitution experiments, each of the three N-hydroxyimide oxygen was found to be essential for inhibition, and derivatization of the hydroxyl group was not tolerated (e.g., compounds 2, 3) (Klumpp and Mirzadegan 2006). However, select substitutions on the phenyl ring were found to increase the potency of N-hydroxyimide inhibitors (compound 4), perhaps via interaction with amino acid residues at the periphery of the RNase H active site.
NMR and fluorescence quenching analysis using the isolated HIV-1 RNase H domain established that the binding of N-hydroxyimides to the domain was metal-ion dependent (Hang et al. 2004). Moreover, the inhibitory potencies of these compounds against RT-associated and isolated RNases H were similar, validating the notion that the N-hydroxyimides bind at the RNase H active site. In buffered aqueous solution, metal ions were found to shift the structural equilibrium of N-hydroxyimides toward the tautomeric enol form, although the affinity of these compounds for free Mg+2 was low. It is likely that the enol form is also assumed when inhibitors bind metal ions at the RNase H active site. The nature of inhibitor binding was confirmed in high-resolution crystal structures of the isolated RNase H domain together with Mn+2 ions and N-hydroxyimide compounds (Klumpp and Mirzadegan 2006). In these studies, co-crystallization was found to be metal-ion dependent, and the resulting structure showed the two metal ions in coordination with the hydroxyimide three-oxygen pharmacophore. In addition, the flexible “His loop,” which is unresolved in some unliganded RNase H structures, appeared to be stabilized by direct interaction with the inhibitor.
Although coordinates for the HIV-1 RNase H-N-hydroxyimide co-crystal structure are not publicly available, a related active site inhibitor N-hydroxy quinazolinedione (compound 5) has been co-crystallized with Mn+2 and an isolated HIV-1 RNase H domain containing the basic turn helix motif of E. coli RNase H (Fig. 7.5a) (Lansdon et al. 2011). Arrangement of the three-oxygen pharmacophore in N-hydroxy quinazolinedione exactly matches that of hydroxyimides; however, a nitrogen-for-carbon substitution in the former compound creates a bicyclic aromatic scaffold that is more stable in aqueous solution (Kirschberg et al. 2009). While coordination of Mn+2 ions by 5 closely resembles that observed with hydroxyimides and other active site inhibitors, the distal phenyl ring assumes an additional edge-on π-stacking interaction with the imidazole side chain of H539. This interaction likely serves to increase the binding affinity of this inhibitor, as well as stabilize the flexible “His loop” in a position close to the active site metal ions.
Metal-binding inhibitors bound at the HIV-1 RNase H active site. Mn+2 ions are depicted as blue-white spheres, while inhibitors and “His loop” residues H539 and A538 are colored according to their atomic constituents. (a) 5 (N-hydroxy quinazolinedione) both coordinates active site metal ions and contributes to an edge-on π-stacking interaction with H539 (Lansdon et al. 2011). The hydroxylated tropolones (b) 10 (β-thujaplicinol) (Himmel et al. 2009) and (c) 11 (manicol) (Chung et al. 2011) likewise bind metal ions and participate in hydrogen bonding/ionic and edge-to-face π-stacking interactions with H539, respectively. An ionic interaction between the imidazole side chain H539 and a manicol tropylium ion has also been proposed. (d) The distal ring in 15, a pyrimidinol carboxylic acid, may also interact with H539 by means of an edge-to-face π-stacking interaction (Kirschberg et al. 2009)
4.1.2 Diketoacids
An alternative form of the three-oxygen pharmacophore targeting two metal-ion active sites can be found in diketoacids. These compounds have been described as inhibitors of polynucleotide transferases such as influenza endonuclease, HCV polymerase, and HIV-1 IN as well as HIV-1 RNase H (Hazuda et al. 2000; Shaw-Reid et al. 2003; Summa et al. 2004; Tomassini et al. 1994). The first diketoacid reported to inhibit HIV-1 RNase H (IC50 = 3.2 μM) was 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid (BTDBA, compound 6) (Shaw-Reid et al. 2003). Although this and similar diketoacids have also been shown to inhibit HIV-1 IN, BTDBA is clearly a selective inhibitor, with reported IC50 values against E. coli RNase H and the polymerase activity of HIV-1 RT both exceeding 50 μM. As with N-hydroxyimides, there is substantial evidence that BTDBA inhibits RNase H by binding to metal ions at the RNase H active site. IC50s of BTDBA against (i) an isolated, recombinant RNase H domain containing the E. coli RNase H basic loop-helix motif and (ii) RT containing a D185N polymerase active site mutation were 4.7 and 8.8 μM, respectively, indicating that RNase H inhibition by BTDBA is polymerase domain independent (Shaw-Reid et al. 2003). Moreover, isothermal calorimetric studies demonstrated that inhibitor binding is metal-ion dependent and does not require the presence of substrate. In in vitro strand transfer activity assays, BTDBA acted synergistically with both NRTIs and NNRTIs, confirming that strand transfer requires both activities and that the inhibitory mechanisms of these compounds are independent and mutually compatible (Shaw-Reid et al. 2005). Despite its in vitro potency, however, BTDBA did not inhibit virus replication in cell culture.
RDS1643 (compound 7), a diketoacid ethyl ester, was also demonstrated to be a selective inhibitor of HIV-1 RNase H, with IC50 values of 13, >100, and >100 μM against RT-associated RNase H, E. coli RNase H, and RT polymerase activities, respectively (Tramontano et al. 2005). However, this inhibitor is notably less potent than BTDBA and like compounds and only weakly inhibited avian myeloblastosis virus (AMV) RT-associated RNase H and HIV-1 IN activities (IC50 = 92 μM and 98 μM, respectively). This is likely due to the masking of the carboxylic acid group with an ethyl ester, a derivatization shown to reduce or destroy the inhibitory activity of diketoacid inhibitors of influenza endonuclease (Tomassini et al. 1994). However, perhaps owing to this ester modification, RDS1643 was found to have antiviral activity in MT-4 cell culture (EC50 = 14 μM). Viruses containing various NNRTI resistance mutations in the RT gene were similarly susceptible, indicating that the mechanisms of inhibition differ between these two types of inhibitors.
Kinetic analysis demonstrated that RDS1643 inhibits RNase H activity via a classical noncompetitive mechanism (Tramontano et al. 2005). Moreover, biochemical studies showed that inhibitor binding to the HIV-1 RNase H domain is reversible and does not require substrate but does require divalent metal ions. These results suggest that, like BTDBA, RDS1643 coordinates the tandem metal ions at the RNase H active center. However, the superior antiviral potency of RDS1643 compared to true diketoacids may be explained by greater cellular permeability due to charge neutralization of the carboxylic acid moiety. Potential intracellular removal of the ester group by cellular esterases might then restore the pharmacophore and convert RDS1643 into a more active compound inside the cell.
Other diketoacid compounds demonstrated to inhibit influenza endonuclease and HIV-1 IN in vitro and/or in vivo were shown to be inactive against HIV-1 RNase H (Parkes et al. 2003; Tomassini et al. 1994) (compounds 8, 9). To understand the structural basis for this specificity, these compounds, along with BTDBA and RDS1643, were superimposed upon the HIV-1 RNase H-Mn+2-hydroxyimide co-crystal structure by molecular modeling (Klumpp and Mirzadegan 2006). Interestingly, for those compounds that were inactive against HIV-1 RNase H, portions of the inhibitor outside of the three-oxygen pharmacophore were predicted to be in steric clash with P537 and/or A538 of the flexible “His loop.” This suggests that the “His loop” motif may play an important role in determining inhibitor specificity and potency while providing a potential secondary target for design of next-generation RNase H active site inhibitors.
4.1.3 Hydroxylated Tropolones
Using a fluorescence-based assay system (Parniak et al. 2003; Shaw-Reid et al. 2005) to screen a large natural product library, β-thujaplicinol was identified as a potent and selective inhibitor of HIV-1 RNase H (IC50 = 0.21 μM) (Budihas et al. 2005). Derived from the plant Thuja plicata, β-thujaplicinol is an α-hydroxytropolone and, as such, possesses a three-oxygen pharmacophore similar to those of hydroxyimides and diketoacids. β-Thujaplicinol is also active against HIV-2 RNase H, which precludes binding of the inhibitor to the NNRTI binding site, but is only weakly active or inactive against E. coli and human RNases H, as well as the polymerase activity of HIV-1 RT. Despite its in vitro potency and selectivity, β-thujaplicinol lacks antiviral activity in cell culture.
The mechanism of β-thujaplicinol-mediated inhibition of HIV-1 RNase H activity has been investigated in detail. One study examines the effects of the inhibitor on stabilized RT-RNA/DNA complexes, as well as in experiments in which the order of addition of enzymatic reaction components is varied (Beilhartz et al. 2009). In HIV-1 RT-RNA/DNA complexes stabilized in either a “pre-” or “post-translocation” complex, β-thujaplicinol did not inhibit RNase H cleavage, suggesting that it can neither bind RT nor inhibit RT-associated RNase H when the RNase H active center is occupied by substrate. In order-of-addition experiments, no inhibition of RNase H activity was observed unless RT, Mg+2, and inhibitor were pre-incubated and the reaction initiated by addition of substrate. Moreover, cleavage reactions, although slowed, eventually ran to completion, regardless of the order in which the reaction components were added. Taken together, these data strongly suggest that β-thujaplicinol (i) inhibits HIV-1 RNase H by binding the Mg ions at the RNase H active site, (ii) competes with substrate for active site occupancy, and (iii) will eventually be displaced by substrate over the course of the reaction.
This apparently competitive mode of inhibition differs from previous reports of β-thujaplicinol and other active site inhibitors behaving noncompetitively (i.e., are able to bind enzyme in the absence or presence of substrate) (Budihas et al. 2005; Tramontano et al. 2005). To reconcile this apparent discrepancy, it has been suggested that β-thujaplicinol selectively inhibits secondary RNase H cleavage and/or binds the enzyme-product complex once the primary cleavage is complete. Moreover, independent biochemical data suggest that an RNA/DNA hybrid can associate with a preformed RT-β-thujaplicinol complex, even if the reverse is not possible (Beilhartz et al. 2009; Budihas et al. 2005; Himmel et al. 2009). These findings are consistent with both classical noncompetitive kinetics and the observation that β-thujaplicinol cannot bind the enzyme-substrate complex.
Crystal structures of either HIV-1 RT or the isolated RNase domain in complex with β-thujaplicinol confirm that the inhibitor binds at the RNase H active site via metal-ion coordination (Fig. 7.5b) (Himmel et al. 2009). As was the case with the N-hydroxyimide and compound 5, β-thujaplicinol forms secondary contacts with the flexible “His loop,” thereby stabilizing it in a position near the RNase H active site. The side chain of R557 is also in close proximity to bound β-thujaplicinol, albeit on the side opposite that of the “His loop.” Superimposition of RT-β-thujaplicinol and human RNase H1-RNA/DNA co-crystal structures (Himmel et al. 2009; Nowotny et al. 2007) predicts that β-thujaplicinol binding would be sterically incompatible with both the scissile phosphate in the RNA strand and the water molecule positioned for nucleophilic attack. Consequently, it appears that a molecular complex containing HIV-1 RT, an RNA/DNA hybrid, and β-thujaplicinol could only exist if the trajectory of the hybrid was altered so that it was accommodated normally, or nearly so, in the template-primer binding cleft, but was displaced relative to the RNase H active site.
Another hydroxytropolone, manicol (compound 11), is structurally similar to β-thujaplicinol except for a bulky aliphatic substituent on the tropolone ring. Manicol exhibits a high degree of inhibitory potency against HIV-1 RT-associated RNase H in vitro (IC50 = 0.6 μM) but, like β-thujaplicinol, has no antiviral activity in cell culture (Chung et al. 2011). In an HIV RT co-crystal structure, TMC278 (rilpivirine), an NNRTI, is bound in the NNRTI binding site, while manicol coordinates Mn+2 in the RNase H active site (Chung et al. 2011) (Fig. 7.5c). Although the aliphatic substituent of manicol generally projects away from the RNase H active site, multiple carbon atoms are within 4 Å of the H539 imidazole side chain in the stabilized “His loop.” Accordingly, the 2-isopropanyl group at the apex of the aliphatic substituent was selected for derivatization for the purpose of increasing the number of secondary enzyme-inhibitor contacts and improving the binding/efficacy of the inhibitor.
While differences in inhibitor potency among manicol and manicol derivatives were modest, several derivatives displayed antiviral activity in cell culture – the most notable being compound 12 (Chung et al. 2011). The mechanistic basis for this improved antiviral function is unclear, although increased hydrophobicity resulting in improved cellular entry may have been instrumental. Compounds 13 and 14 also inhibited the DNA polymerase activity of HIV-1 RTs containing either an E478Q or a D549A mutation. Since both of these mutations preclude binding of one or both metals at the RNase H active site and binding of hydroxytropolones to the active site is metal-ion dependent, it is possible that inhibition of DNA synthesis by these manicol derivatives is the result of binding to an unknown second site within RT.
4.1.4 Pyrimidinol Carboxylic Acids
A fourth scaffold for the metal-binding three-oxygen pharmacophore can be found in pyrimidinol carboxylic acid derivatives (Kirschberg et al. 2009). Several of these compounds are active against HIV-1 RT-associated RNase H, with IC50s in the low to submicromolar range, and are only weakly active or inactive against human RNase H1. One derivative (compound 15) was successfully crystallized together with Mn+2 and the isolated RNase H domain of HIV-1 RT containing the basic loop-helix motif of E. coli RNase H (Fig. 7.5d). In this structure, resolved to 1.7 Å, the carboxyl group of the inhibitor is shown to coordinate metal ion B, while metal ion A is coordinated by two phenolic oxygen atoms. Moreover, C2 in the phenyl substituent interacts directly with imidazole side chain of H539, possibly through an edge-to-face π-π interaction. None of the pyrimidinol carboxylic acid derivatives possess antiviral activity in cell culture.
4.2 Allosteric Inhibitors
Allosteric inhibition of RNase H activity is an attractive alternative to active site inhibitors. Allosteric inhibitors need not contain the negatively charged three-oxygen pharmacophore necessary for metal-ion coordination and therefore have potential for greater cell permeability. Moreover, binding of such inhibitors is not limited to the face of the active site, or even the RNase H domain, so sites more amenable to high-affinity interaction with small molecules can be utilized.
As previously noted, NNRTIs are excellent examples of allosteric inhibitors. Originally identified as potent inhibitors of the HIV-1 RT polymerase activity, NNRTIs bind near the base of the p66 thumb subdomain 10–15 Å from the DNA polymerase active site (K. Das et al. 2012; Kohlstaedt et al. 1992). This binding site is created by an induced fit mechanism in the presence of inhibitor and is not apparent otherwise. NNRTIs have also been shown to modulate RT-associated RNase H activity, indicating that inhibitor binding may induce global structural changes affecting positioning of the RNase H domain, substrate binding, or both. Two other classes of allosteric RNase H inhibitors have been identified: N-acyl hydrazones and vinylogous ureas, the latter of which affect RNase H activity almost exclusively (Fig. 7.6).
4.2.1 N-Acyl Hydrazones
N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazine (BBNH, compound 16) is an N-acyl hydrazone shown to inhibit HIV-1 RT polymerase and RNase H activities with equal efficiency (IC50s range from 0.8 to 15 μM, depending on the assay) (Borkow et al. 1997). The compound also inhibits E. coli and Moloney murine leukemia virus (Mo-MLV) RT-associated RNases H (IC50s = 2.7 and 0.8 μM), but not the polymerase and RNase H activities of HIV-2 RT (IC50s > 50 μM). BBNH displays moderate antiviral activity in MT4 cell culture (IC50 = 5 μM) but is cytostatic or cytotoxic at concentrations above 10 or 25 μM, respectively.
Inhibition of HIV-1 RNase H by BBNH requires pre-incubation of RT and inhibitor, is reversible, and is linearly competitive with nucleic acid substrate (Borkow et al. 1997). In an experiment designed to explore potential interaction(s) between BBNH and NNRTIs, BBNH was found to inhibit inactivation of HIV-1 RT polymerase function by a photoactivatable analog of nevirapine. With respect to RNase H activity, however, the potency of BBNH was not diminished in the presence of NNRTIs UC38 and TIBO nor was it affected after photo-cross-linking a nevirapine derivative to the catalytic RT. Moreover, BBNH inhibited the polymerase and RNase H activities of RTs containing any of a number of NNRTI resistance mutations (K103N, Y181I, Y188H, or Y188L) to approximately the same degree as the wild-type enzyme – with one exception. Although BBNH inhibited the polymerase activities of wild-type and Y181C RT with equal potency, the inhibitor was completely ineffective against the RNase H activity of the mutant enzyme (IC50 > 50 μM).
Taken together, these data suggested that the BBNH binds RT at a site different from, but perhaps overlapping, the NNRTI binding site. Certainly, NNRTI binding and/or activity is affected by BBNH, although it is not clear that the reverse is true. It was further proposed that BBNH-mediated inhibition of RNase H activity is caused by binding of the inhibitor to a second site closer to or within the RNase H domain (Borkow et al. 1997). This notion is supported by the apparent independence of BBNH and NNRTI function with respect to inhibition of RNase H activity, as well as the observation that BBNH inhibits E. coli RNase H, an enzyme with little homology to the polymerase domain or NNRTI binding site within HIV-1 RT.
A molecular model of the putative second binding site was generated in which the hydrazone moiety of BBNH was proposed to interact with the metal ions at the RNase H active site, while the distal aromatic ring of the inhibitor participated in a stacking interaction with the side chain of RNase H primer grip residue Y501 (Arion et al. 2002). To test this model, HIV-1 RTs containing a series of Y501 substitution mutations were expressed, purified, and assayed for enzyme activity in the presence and absence of inhibitor. In large part, the study supported involvement of Y501 in inhibitor binding. Although the RNase H activity of Y501F RT remained sensitive to BBNH, RT containing a Y501R mutation, which would likely be less amenable to π stacking, was completely resistant.
The activity profiles of BBNH analogs were in some cases shown to be dramatically different from those of the parent compound (Arion et al. 2002; Klumpp and Mirzadegan 2006; Sluis-Cremer et al. 2002). For example, BBSH (compound 17), which contains a phenol group in place of the naphthol moiety, inhibited HIV-1 RT polymerase activity (IC50 = 2.5 μM) but not RNase H (IC50 > 50 μM). Conversely, IC50s for DABNH (compound 18) against RT-associated polymerase and RNase H activities were >40 and 4 μM, respectively. The effects of these compounds on the stability of the HIV-1 RT heterodimer were also variable. Specifically, BBNH and BBSH reduced the stability of the HIV-1 RT heterodimer, while DABNH appeared to have no effect (Sluis-Cremer et al. 2002). Clearly, the inhibitory profiles of N-acyl hydrazones can be greatly affected by relatively small structural changes.
The chlorophenylhydrazone of mesoxalic acid (CPHM, compound 19) was identified as an inhibitor of strand transfer catalyzed by HIV-1 RT (Gabbara et al. 1999). This compound inhibits strand transfer, polymerase, and RNase H activities with approximately equal potency (IC50s = 4.5, 2.3, and 3.0 μM, respectively) but possesses no antiviral activity. Interestingly, CPHM did not inhibit the RNase H activity of RT containing a polymerase active site mutation (D185N), while in the reverse experiment, inhibition of RT polymerase activity was unaffected by mutation of the RNase H active site (D443N) (Shaw-Reid et al. 2003). These data suggest the CPHM inhibits both RT activities by binding in the polymerase domain, though perhaps at a site(s) different from that recognized by BBNH. The latter supposition is supported by the markedly different chemical composition of CPHM compared to BBNH, BBSH, and DABNH, which are all relatively similar by comparison (Fig. 7.6).
Another hydrazone (E)-3,4-dihydroxy-N′-((2-methoxynaphthalen-1-yl)methylene)benzohydrazide (DHBNH, compound 20) potently inhibits HIV-1 RT-associated RNase H (IC50 = 0.5 μM)and is inactive against RT polymerase activity (Himmel et al. 2006). DHBNH also possesses antiviral activity (IC50 4.5 μM), with an improved selectivity index relative to BBNH (CC50 > 100 μM). A co-crystal structure with HIV-1 RT shows DHBNH bound among residues D186, Y188, and W229 of the p66 subunit, approximately 15 Å from the NNRTI binding site and ~50 Å from the RNase H active site (Fig. 7.7). No association between DHBNH and either the RNase H domain or metal ions was observed. Moreover, DHBNH only weakly inhibits RNase H activity catalyzed by the isolated HIV-1 RNase H domain (Himmel et al. 2006), suggesting that its inhibitory mechanism is truly allosteric.
Allosteric inhibitors bound to HIV-1 RT. (a) 20 (DHBNH) binds to the inner surface of the p66 palm subdomain near the base of the p66 thumb (Himmel et al. 2006). The p51 subunit (gray) as well as the fingers (blue), palm (magenta), and thumb subdomains of p66 are highlighted. (b) A closer view of the 20 (DHBNH) binding site. Primer grip residues comprising or adjacent to the binding site are highlighted. (c) The NNRTI rilpivirine is enveloped within the p66 palm subdomain at the base of the p66 thumb subdomain (Das et al. 2008). In contrast to DHBNH, bound rilpivirine is not directly exposed to the nucleic acid-binding cleft. Instead, this inhibitor accesses its binding site from the opposite side of the enzyme. (d) A closer view of rilpivirine binding. Note that in both (c) and (d), it was necessary to remove p66 residues 227–234 in order to reveal the NNRTI binding site
It is difficult to reconcile the single DHBNH binding site revealed in the co-crystal structure with either exclusive inhibition of HIV-1 RNase H activity or the two-binding-site model proposed for BBNH. One explanation is that crystal packing prevented DHBNH binding to the second site or that binding to the second site is somehow substrate dependent. Alternatively, DHBNH and other N-acyl hydrazones may bind only to the site identified in the crystal structure, affecting subtle changes in RT structure and/or flexibility that alter the way the substrate is accommodated by the enzyme (Himmel et al. 2006). These changes might then manifest themselves in either reduced polymerase or RNase H activity or both, depending on the structure of the inhibitor, its effect on RT, and the nature of the perturbation in substrate binding. Regardless of which hypothesis is proven correct, it is clear that additional study is required to elucidate the mechanism(s) of action of this complex class of inhibitors.
4.2.2 Vinylogous Ureas and Thienopyrimidinones
The lead vinylogous urea compounds were identified in a high-throughput robotic screen of ~230,000 natural and synthetic compounds from a series of National Cancer Institute libraries. 2-Amino-5,6,7,8-tetrahydro-4H-cyclohepta[b]thiophene-3-carboxamide (NSC727447, compound 21) and N-[3-(aminocarbonyl)-4,5-dimethyl-2-thienyl]-2-furancarboxamide (NSC747448, compound 22) were shown to potently inhibit HIV-1 and HIV-2 RNases H, with IC50s of 2.0 and 3.2 μM, respectively, but only weakly inhibited HIV-1 replication in cell culture (Wendeler et al. 2008). Both inhibitors were also active against human RNase H1 to a moderate degree (IC50s = 10.6 μM and 29 μM for NSC727447 and NSC727448, respectively), although neither appreciably inhibited E. coli RNase H activity (IC50s >100 and 73 μM). Yonetani-Theorell analysis demonstrated that while the inhibitory activities of NSC727447 and β-thujaplicinol are mutually exclusive, only the vinylogous ureas bind RT in the presence of substrate. These data suggest that vinylogous ureas and RNase H active site inhibitors differ in their inhibitory mechanisms.
Structure-activity relationship analysis of vinylogous ureas and their cyclized counterparts, thienopyrimidinones, demonstrated that in addition to the 2-amino-thiophene-3-carboxamide core shared by NSC727447 and NSC727448, peripheral functional groups were important for inhibition of RNase H activity as well (Chung et al. 2010). For example, removing or changing the size of the heptane ring in NSC727447 was extremely detrimental to inhibitory function, as was substituting the furan group of NSC727448 with a 3-methyl-4-ethyl-chlorobenzene moiety. However, one thienopyrimidinone, NSC727665 (compound 23), was found to be more active than either of the lead compounds, with an IC50 of 0.85 μM against HIV-1 RT-associated RNase H. In this compound, the nitro group was found to be essential, as derivatives in which the group was either removed or replaced with a methoxy group were completely devoid of inhibitory function.
Mass spectrometric and mutational studies suggest that the vinylogous ureas bind near the p51-RNase H domain interface at the base of the p51 thumb (Wendeler et al. 2008). Molecular docking of NSC727447 and NSC727665 into this site suggests that the binding pocket is formed by p51 residues K275, V276, C280, and R284, among others, as well as p66 residues G541 and H539 (Chung et al. 2010). The potential involvement of H539 suggests a mechanism of inhibition that may involve interference with the function of the flexible, catalytically important “His loop.” Alternatively, because the putative binding site is located at the p66-p51 interface, it is possible that binding of vinylogous ureas and/or thienopyrimidinones affects dimer stability. Scanning mutagenesis of residues K275 through T286 of the p51 thumb subdomain indicates that C280 and T286 are important for binding of NSC727665, as mutation of these residues produced enzymes that were resistant to RNase H inhibition (Chung et al. 2012). Interestingly, these mutations did not affect inhibitory susceptibility to NSC727447, suggesting that this inhibitor may bind to an alternative site or form different contacts with the enzyme.
4.3 RNase H Inhibitors with Unknown Binding Sites
For a subset of RNase H inhibitors, binding sites and mechanisms of action remain unknown. These include quinones and naphthoquinones, nucleotides and dinucleotides, mappicine analogs, thiocarbamates, and triazoles (Fig. 7.8).
4.3.1 Quinones and Naphthoquinones
Illimaquinone (compound 24), a marine sponge metabolite, is one of the first RNase H inhibitors to be identified. This compound weakly inhibits HIV-1-, HIV-2-, and E. coli-associated RNases H, with IC50 values in the 15–50 μM range, and is virtually inactive against the polymerase activities of HIV-1 and HIV-2 RT (Loya et al. 1990; Min et al. 2002). In contrast, the related compounds 6′-hydroxy avarol (compound 25) and 6′-hydroxy-4-methoxy avarone (compound 26) inhibit both activities of HIV-1 RT with similar potency (Loya and Hizi 1990).
Limited structure-activity relationship studies leave the mechanism(s) of action of this class of inhibitors unclear. Among naphthoquinone derivatives, the unsubstituted compound (compound 27) was the most potent against HIV-1 RT-associated RNase H activity, while hydroxylated derivatives (e.g., compound 28) demonstrated both increased potency against polymerase activity and reduced effectiveness against RNase H (Min et al. 2002). Illimaquinone was shown to be antagonistic with N-ethylmaleimide (NEM), a reagent shown to inhibit RT-associated DNA polymerase activity by covalently modifying residue C280 (Loya and Hizi 1993). This suggests an inhibitory mechanism involving binding to or modification of cysteine side chains, a notion supported by the observation that RT containing a C280S mutation is resistant to illimaquinone-mediated inhibition.
4.3.2 Nucleotides and Dinucleotides
The nucleotide analog AZT-MP (compound 29) was demonstrated to weakly inhibit HIV-1 RT-associated RNase H activity, while the di- and triphosphate derivatives of this compound (AZT-DP and AZT-TP) are completely ineffectual as RNase H inhibitors(Tan et al. 1991). Interestingly, the potency and mechanism of action of AZT-MP was shown to be dependent upon both substrate and the metal ion used to catalyze the reaction (Zhan et al. 1994). Specifically, cleavage of poly-rG:dC was inhibited more effectively in the presence of Mn+2 ions (IC50 = 25 μM) than when Mg+2 ions were utilized as the RT cofactor (IC50 = 250 μM). Moreover, kinetic analysis revealed that the mechanisms for inhibiting RNase H in reactions utilizing poly-rG:dC or poly-rA:dT as substrate were uncompetitive/mixed or competitive, respectively. The AZT-MP binding site for inhibition of RT-associated RNase H activity remains unresolved.
Among unmodified nucleotide dimers, diguanosine (rGrG) was found to be the most potent against HIV-1 RT-associated RNase H activity (IC50 = 15 μM) (Allen et al. 1996). However, this dinucleotide did not inhibit RT-associated polymerase activity nor did it inhibit either E. coli RNase H1 or RNase H activity in HeLa cell nuclear extracts. Although not a particularly robust inhibitor, the potency of diguanosine against RT-associated RNase H could be improved by substitution of the 5′-3′ diester linkage with a 5′-2′-formacetal (IC50 = 5 μM) (compound 30).
4.3.3 Mappicine Analogs, Thiocarbamates, and Triazoles
Select mappicine analogs (e.g., compounds 31–33), thiocarbamates (e.g., compound 34), and triazoles (e.g., compound 35) have been described as selective HIV-1 RNase H inhibitors, some of which also possess antiviral activity in cell culture (Curran et al. 2004; Di Grandi et al. 2010). In the case of mappicine analogs, for example, antiviral IC50 values ranging from 2.1 to 10 μM have been reported. In addition, virus containing NNRTI resistance mutations (i.e., K103N/Y181C or Y106A/Y181C) remained susceptible to mappicine analogs, suggesting that these RNase H inhibitors do not bind at the NNRTI binding site.
Since none of these compounds contain substituents that resemble the three-oxygen pharmacophores common among RNase H active site inhibitors, it is likely that they act allosterically. However, the actual binding sites and mechanisms of action of these inhibitors remain unknown.
5 RNase H and DNA Polymerase Inhibitor Interaction
Highly active antiretroviral therapy, or HAART, refers to multidrug regimens that target multiple HIV proteins/functions simultaneously (Asahchop et al. 2012). Because HAART is currently the best clinical options for controlling HIV infection, it is important to determine whether inhibitors developed to specifically target HIV RT-associated RNase H are also compatible with the NRTI and NNRTI components of these regimens.
While there is no evidence to suggest that NRTIs affect the function of RNase H inhibitors or vice versa, it has been speculated that inhibition of RNase H activity by any means may decrease the sensitivity of RT to the thymidine analog AZT (azidothymidine, zidovudine) (Delviks-Frankenberry et al. 2007; Ehteshami et al. 2008; Nikolenko et al. 2007). Sequencing of the RT gene in AZT-resistant clinical isolates revealed a common set of mutations proximal to the polymerase active site. These mutations, collectively referred to as thymidine analog mutations, or TAMs, were determined to be sufficient to confer AZT resistance to RT by increasing the rate at which the enzyme catalyzed AZT excision. Other mutations (e.g., N348I, A360V, and D549N) within the connection subdomain and RNase H domain of the enzyme were subsequently found to enhance AZT resistance in these enzymes, although the mechanism by which this occurred was unclear. It was speculated that the mutations in the connection subdomain and RNase H domain reduced RNase H activity in the AZT-resistant enzymes and that doing so prolonged the residency time of RT on the RNA/DNA substrate when minus-strand synthesis was prematurely terminated by incorporation of AZT. This, in turn, would allow more time for RT to excise the chain terminator and resume DNA synthesis (Delviks-Frankenberry et al. 2010).
Kinetic analysis of recombinant enzymes containing the connection and RNase H domain mutations partially confirmed this hypothesis. Several of these mutations did, in fact, reduce RNase H activity, and this reduction generally correlated with increased AZT resistance independent of TAMs (Delviks-Frankenberry et al. 2008). Some connection subdomain mutations, however, appeared to directly affect the rate of AZT excision, possibly by affecting how the RNA/DNA substrate is positioned within the template-primer binding cleft. The relative importance of these RNase H-dependent and RNase H-independent mechanisms of enhancing AZT resistance remains to be determined, as does the potential for antagonism between RNase H inhibitors and AZT. In a recent study, it was determined that four of five RNase H inhibitors tested had little effect on the AZT susceptibility of HIV in cell culture, while the fifth decreased susceptibility approximately fivefold (Davis et al. 2011). None of the RNase H inhibitors had any effect on HIV susceptibility to another NRTI, the cytidine analog 3TC (lamivudine).
It is worth noting that resistance to NRTIs other than AZT is achieved not by increasing the rate at which chain terminators are excised from nascent DNA but rather by active site discrimination, which reduces the frequency with which nucleoside analogs are incorporated (Sluis-Cremer et al. 2000). Consequently, impairing RNase H function by inhibition or mutation should have no effect on resistance to these inhibitors, given that there would be no obvious advantage to increasing residency time on a chain-terminated substrate. This notion is confirmed in experiments with d4T (stavudine)-resistant RT, where introduction of N348I and A360V connection subdomain mutations had no effect on inhibitor susceptibility (Ehteshami et al. 2008).
Studies have shown that interactions between NNRTIs and active site-binding RNase H inhibitors vary with the compounds being evaluated, the activity being measured, and the assay being utilized. In addition to the aforementioned mixed interactions between NNRTIs and the acyl hydrazone BBNH (Borkow et al. 1997), efavirenz and diketoacid analogs were found to behave antagonistically with respect to RNase H inhibition, additively in an RNA-dependent DNA synthesis assay, and synergistically in an assay system requiring the coordinated completion of multiple stages of reverse transcription (Shaw-Reid et al. 2005). In contrast, another NNRTI, calanolide A, and β-thujaplicinol were shown to behave synergistically with respect to RNase H inhibition (Budihas et al. 2005). These mixed results are perhaps not surprising, given the allosteric mechanism of NNRTIs and their capacity to either agonize or antagonize RNase H function, depending on the compound. Moreover, the inhibitory potencies of NNRTIs have been shown to be partially dependent upon substrate sequence and structure (Hang et al. 2007), rendering potential interactions between NNRTIs and RNase H inhibitors highly context dependent.
6 Conclusions and Perspectives
Success in developing potent and specific RNase H inhibitors has been mixed. Active site inhibitors have been developed with submicromolar IC50s against HIV-1 RT-associated RNase H, and most are only weakly active against RT polymerase and E. coli RNase H activities. Several X-ray co-crystal structures have elucidated how and where these inhibitors bind in the RNase H active center, as well as how secondary contacts with the “His loop” might be exploited in future rounds of derivatization. However, most of the current RNase H active site inhibitors have low selectivity indices, most likely due to poor cellular entry and/or cytotoxicity, and possess minimal to moderate antiviral activity.
Because they do not require the three-oxygen pharmacophore necessary for coordination of tandem metal ions and can theoretically bind any site on the surface of HIV-1 RT, allosteric RNase H inhibitors perhaps offer greater promise as therapeutic agents. This potential is further accentuated by the observation that some compounds (NNRTIs, BBNH) inhibit the polymerase and RNase H activities of RT simultaneously. Both N-acyl hydrazones and thienopyrimidinones have been shown to possess strong to moderate antiviral activities in cell culture; however, no member of either class of compounds has as yet been proven sufficiently effective to be entered into clinical trials.
Inclusion of both an IN active site inhibitor and NNRTIs in current HAART regimens offers hope that comparable active site and allosteric RNase H inhibitors may be developed. Engineering RNase H active site inhibitors to more actively engage the flexible “His loop,” or even select bases within the RNA substrate, could increase both the potency and selectivity of these compounds. Pharmacokinetics might also be improved by synthesizing charge-neutral, inert derivatives of active site inhibitors that might be activated by cellular enzymes after passing through the plasma membrane. With respect to allosteric RNase H inhibitors, identifying the binding sites and determining the mechanisms of action of thienopyrimidinones, mappicine analogs, and other inhibitory compounds is an important first step toward improving their efficacy. Moreover, the development of fluorescent assay systems for measuring RT-associated RNase H activity will undoubtedly accelerate identification of new RNase H inhibitors.
Abbreviations
- ddNTP:
-
Dideoxynucleoside triphosphate
- dNTP:
-
Deoxynucleoside triphosphate
- ds:
-
Double stranded
- HIV-1:
-
Human immunodeficiency virus type 1
- HIV-2:
-
Human immunodeficiency virus type 2
- IN:
-
Integrase
- Mo-MLV:
-
Moloney murine leukemia virus
- NNRTI:
-
Non-nucleoside RT inhibitor
- NRTI:
-
Nucleoside RT inhibitor
- nt:
-
Nucleotide
- PPT:
-
Polypurine tract
- PR:
-
Protease
- RNase H:
-
Ribonuclease H
- RT:
-
Reverse transcriptase
References
Allen SJW, Krawczyk SH, McGee LR, Bischofberger N, Mulato AS, Cherrington JM (1996) Inhibition of HIV-1 RNase H activity by nucleotide dimers and monomers. Antivir Chem Chemother 7(1):37–45
Arion D, Sluis-Cremer N, Min KL, Abram ME, Fletcher RS, Parniak MA (2002) Mutational analysis of Tyr-501 of HIV-1 reverse transcriptase. Effects on ribonuclease H activity and inhibition of this activity by N-acylhydrazones. J Biol Chem 277(2):1370–1374
Asahchop EL, Wainberg MA, Sloan RD, Tremblay CL (2012) Antiviral drug resistance and the need for development of new HIV-1 reverse transcriptase inhibitors. Antimicrob Agents Chemother. doi:10.1128/AAC.00591-12
Beilhartz GL, Wendeler M, Baichoo N, Rausch J, Le Grice S, Gotte M (2009) HIV-1 reverse transcriptase can simultaneously engage its DNA/RNA substrate at both DNA polymerase and RNase H active sites: implications for RNase H inhibition. J Mol Biol 388(3):462–474
Borkow G, Fletcher RS, Barnard J, Arion D, Motakis D, Dmitrienko GI, Parniak MA (1997) Inhibition of the ribonuclease H and DNA polymerase activities of HIV-1 reverse transcriptase by N-(4-tert-butylbenzoyl)-2-hydroxy-1-naphthaldehyde hydrazone. Biochemistry 36(11): 3179–3185
Budihas SR, Gorshkova I, Gaidamakov S, Wamiru A, Bona MK, Parniak MA et al (2005) Selective inhibition of HIV-1 reverse transcriptase-associated ribonuclease H activity by hydroxylated tropolones. Nucleic Acids Res 33(4):1249–1256
Champoux JJ, Schultz SJ (2009) Ribonuclease H: properties, substrate specificity and roles in retroviral reverse transcription. FEBS J 276(6):1506–1516. doi:10.1111/j.1742-4658.2009.06909.x
Chung S, Wendeler M, Rausch JW, Beilhartz G, Gotte M, O’Keefe BR et al (2010) Structure-activity analysis of vinylogous urea inhibitors of human immunodeficiency virus-encoded ribonuclease H. Antimicrob Agents Chemother 54(9):3913–3921. doi:10.1128/AAC.00434-10
Chung S, Himmel DM, Jiang JK, Wojtak K, Bauman JD, Rausch JW et al (2011) Synthesis, activity, and structural analysis of novel alpha-hydroxytropolone inhibitors of human immunodeficiency virus reverse transcriptase-associated ribonuclease H. J Med Chem 54(13):4462–4473. doi:10.1021/jm2000757
Chung S, Miller JT, Johnson BC, Hughes SH, Le Grice SF (2012) Mutagenesis of human immunodeficiency virus reverse transcriptase p51 subunit defines residues contributing to vinylogous urea inhibition of ribonuclease H activity. J Biol Chem 287(6):4066–4075. doi:10.1074/jbc.M111.314781
Cirino NM, Kalayjian RC, Jentoft JE, Le Grice SF (1993) Fluorimetric analysis of recombinant p15 HIV-1 ribonuclease H. J Biol Chem 268(20):14743–14749
Cowan JA, Ohyama T, Howard K, Rausch JW, Cowan SM, Le Grice SF (2000) Metal-ion stoichiometry of the HIV-1 RT ribonuclease H domain: evidence for two mutually exclusive sites leads to new mechanistic insights on metal-mediated hydrolysis in nucleic acid biochemistry. J Biol Inorg Chem JBIC A Publ Soc Biol Inorg Chem 5(1):67–74
Curran DP, Parniak MA, Gabarda A (2004) USA Patent no. US 2004/0058948 A1. United States Patent Application Publication: U. S. P. Office
Das D, Georgiadis MM (2004) The crystal structure of the monomeric reverse transcriptase from Moloney murine leukemia virus. Structure 12(5):819–829. doi:10.1016/j.str.2004.02.032
Das K, Bauman JD, Clark AD Jr, Frenkel YV, Lewi PJ, Shatkin AJ et al (2008) High-resolution structures of HIV-1 reverse transcriptase/TMC278 complexes: strategic flexibility explains potency against resistance mutations. Proc Natl Acad Sci USA 105(5):1466–1471. doi:10.1073/pnas.0711209105
Das K, Martinez SE, Bauman JD, Arnold E (2012) HIV-1 reverse transcriptase complex with DNA and nevirapine reveals non-nucleoside inhibition mechanism. Nat Struct Mol Biol 19(2):253–259. doi:10.1038/nsmb.2223
Davies JF 2nd, Hostomska Z, Hostomsky Z, Jordan SR, Matthews DA (1991) Crystal structure of the ribonuclease H domain of HIV-1 reverse transcriptase. Science 252(5002):88–95
Davis CA, Parniak MA, Hughes SH (2011) The effects of RNase H inhibitors and nevirapine on the susceptibility of HIV-1 to AZT and 3TC. Virology 419(2):64–71. doi:10.1016/j.virol.2011.08.010
De Clercq E (2009) Anti-HIV drugs: 25 compounds approved within 25 years after the discovery of HIV. Int J Antimicrob Agents 33(4):307–320. doi:10.1016/j.ijantimicag.2008.10.010
Delviks-Frankenberry KA, Nikolenko GN, Barr R, Pathak VK (2007) Mutations in human immunodeficiency virus type 1 RNase H primer grip enhance 3′-azido-3′-deoxythymidine resistance. J Virol 81(13):6837–6845
Delviks-Frankenberry KA, Nikolenko GN, Boyer PL, Hughes SH, Coffin JM, Jere A, Pathak VK (2008) HIV-1 reverse transcriptase connection subdomain mutations reduce template RNA degradation and enhance AZT excision. Proc Natl Acad Sci USA 105(31):10943–10948. doi:10.1073/pnas.0804660105
Delviks-Frankenberry KA, Nikolenko GN, Pathak VK (2010) The “connection” between HIV drug resistance and RNase H. Viruses 2(7):1476–1503. doi:10.3390/v2071476
DeStefano JJ, Bambara RA, Fay PJ (1993) Parameters that influence the binding of human immunodeficiency virus reverse transcriptase to nucleic acid structures. Biochemistry 32(27): 6908–6915
Di Grandi M, Olson M, Prashad AS, Bebernitz G, Luckay A, Mullen S et al (2010) Small molecule inhibitors of HIV RT Ribonuclease H. Bioorg Med Chem Lett 20(1):398–402. doi:10.1016/j.bmcl.2009.10.043
Di Marzo Veronese F, Copeland TD, DeVico AL, Rahman R, Oroszlan S, Gallo RC, Sarngadharan MG (1986) Characterization of highly immunogenic p66/p51 as the reverse transcriptase of HTLV-III/LAV. Science (Washington, DC 1883-) 231(4743):1289–1291
Ehteshami M, Beilhartz GL, Scarth BJ, Tchesnokov EP, McCormick S, Wynhoven B et al (2008) Connection domain mutations N348I and A360V in HIV-1 reverse transcriptase enhance resistance to 3′-azido-3′-deoxythymidine through both RNase H-dependent and -independent mechanisms. J Biol Chem 283(32):22222–22232
Furfine ES, Reardon JE (1991) Reverse transcriptase. RNase H from the human immunodeficiency virus. Relationship of the DNA polymerase and RNA hydrolysis activities. J Biol Chem 266(1):406–412
Gabbara S, Davis WR, Hupe L, Hupe D, Peliska JA (1999) Inhibitors of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. Biochemistry 38(40):13070–13076
Gotte M, Fackler S, Hermann T, Perola E, Cellai L, Gross HJ et al (1995) HIV-1 reverse transcriptase-associated RNase H cleaves RNA/RNA in arrested complexes: implications for the mechanism by which RNase H discriminates between RNA/RNA and RNA/DNA. EMBO J 14(4):833–841
Gotte M, Maier G, Gross HJ, Heumann H (1998) Localization of the active site of HIV-1 reverse transcriptase-associated RNase H domain on a DNA template using site-specific generated hydroxyl radicals. J Biol Chem 273(17):10139–10146
Gotte M, Rausch JW, Marchand B, Sarafianos S, Le Grice SF (2010) Reverse transcriptase in motion: conformational dynamics of enzyme-substrate interactions. Biochim Biophys Acta 1804(5):1202–1212. doi:10.1016/j.bbapap.2009.07.020
Hang JQ, Rajendran S, Yang Y, Li Y, In PW, Overton H et al (2004) Activity of the isolated HIV RNase H domain and specific inhibition by N-hydroxyimides. Biochem Biophys Res Commun 317(2):321–329
Hang JQ, Li Y, Yang Y, Cammack N, Mirzadegan T, Klumpp K (2007) Substrate-dependent inhibition or stimulation of HIV RNase H activity by non-nucleoside reverse transcriptase inhibitors (NNRTIs). Biochem Biophys Res Commun 352(2):341–350
Hansen J, Schulze T, Mellert W, Moelling K (1988) Identification and characterization of HIV-specific RNase H by monoclonal antibody. EMBO J 7(1):239–243
Hare S, Vos AM, Clayton RF, Thuring JW, Cummings MD, Cherepanov P (2010) Molecular mechanisms of retroviral integrase inhibition and the evolution of viral resistance. [Research support, Non-U.S. Gov’t]. Proc Natl Acad Sci USA 107(46):20057–20062. doi:10.1073/pnas.1010246107
Hazuda DJ, Felock P, Witmer M, Wolfe A, Stillmock K, Grobler JA et al (2000) Inhibitors of strand transfer that prevent integration and inhibit HIV-1 replication in cells. Science 287(5453):646–650
Himmel DM, Sarafianos SG, Dharmasena S, Hossain MM, McCoy-Simandle K, Ilina T et al (2006) HIV-1 reverse transcriptase structure with RNase H inhibitor dihydroxy benzoyl naphthyl hydrazone bound at a novel site. ACS Chem Biol 1(11):702–712
Himmel DM, Maegley KA, Pauly TA, Bauman JD, Das K, Dharia C et al (2009) Structure of HIV-1 reverse transcriptase with the inhibitor beta-Thujaplicinol bound at the RNase H active site. Structure 17(12):1625–1635
Huang H, Chopra R, Verdine GL, Harrison SC (1998) Structure of a covalently trapped catalytic complex of HIV-1 reverse transcriptase: implications for drug resistance. Science 282(5394): 1669–1675
Jacobo-Molina A, Ding J, Nanni RG, Clark AD Jr, Lu X, Tantillo C et al (1993) Crystal structure of human immunodeficiency virus type 1 reverse transcriptase complexed with double-stranded DNA at 3.0 A resolution shows bent DNA. Proc Natl Acad Sci USA 90(13):6320–6324
Katayanagi K, Miyagawa M, Matsushima M, Ishikawa M, Kanaya S, Ikehara M et al (1990) Three-dimensional structure of ribonuclease H from E. coli. Nature 347(6290):306–309
Keck JL, Marqusee S (1995) Substitution of a highly basic helix/loop sequence into the RNase H domain of human immunodeficiency virus reverse transcriptase restores its Mn(2+)-dependent RNase H activity. Proc Natl Acad Sci USA 92(7):2740–2744
Keck JL, Goedken ER, Marqusee S (1998) Activation/attenuation model for RNase H. A one-metal mechanism with second-metal inhibition. J Biol Chem 273(51):34128–34133
Kirschberg TA, Balakrishnan M, Squires NH, Barnes T, Brendza KM, Chen X et al (2009) RNase H active site inhibitors of human immunodeficiency virus type 1 reverse transcriptase: design, biochemical activity, and structural information. J Med Chem 52(19):5781–5784. doi:10.1021/jm900597q
Klumpp K, Mirzadegan T (2006) Recent progress in the design of small molecule inhibitors of HIV RNase H. Curr Pharm Des 12(15):1909–1922
Klumpp K, Hang JQ, Rajendran S, Yang Y, Derosier A, Wong Kai In P et al (2003) Two-metal ion mechanism of RNA cleavage by HIV RNase H and mechanism-based design of selective HIV RNase H inhibitors. Nucleic Acids Res 31(23):6852–6859
Kohlstaedt LA, Wang J, Friedman JM, Rice PA, Steitz TA (1992) Crystal structure at 3.5 A resolution of HIV-1 reverse transcriptase complexed with an inhibitor. Science 256(5065): 1783–1790
Lansdon EB, Liu Q, Leavitt SA, Balakrishnan M, Perry JK, Lancaster-Moyer C et al (2011) Structural and binding analysis of pyrimidinol carboxylic acid and N-hydroxy quinazolinedione HIV-1 RNase H inhibitors. Antimicrob Agents Chemother 55(6):2905–2915. doi:10.1128/AAC.01594-10
Loya S, Hizi A (1990) The inhibition of human immunodeficiency virus type 1 reverse transcriptase by avarol and avarone derivatives. FEBS Lett 269(1):131–134
Loya S, Hizi A (1993) The interaction of illimaquinone, a selective inhibitor of the RNase H activity, with the reverse transcriptases of human immunodeficiency and murine leukemia retroviruses. J Biol Chem 268(13):9323–9328
Loya S, Tal R, Kashman Y, Hizi A (1990) Illimaquinone, a selective inhibitor of the RNase H activity of human immunodeficiency virus type 1 reverse transcriptase. Antimicrob Agents Chemother 34(10):2009–2012
Ma JB, Yuan YR, Meister G, Pei Y, Tuschl T, Patel DJ (2005) Structural basis for 5′-end-specific recognition of guide RNA by the A. fulgidus Piwi protein. Nature 434(7033):666–670
Min BS, Miyashiro H, Hattori M (2002) Inhibitory effects of quinones on RNase H activity associated with HIV-1 reverse transcriptase. Phytother Res PTR 16(Suppl 1):S57–S62
Nikolenko GN, Delviks-Frankenberry KA, Palmer S, Maldarelli F, Fivash MJ Jr, Coffin JM, Pathak VK (2007) Mutations in the connection domain of HIV-1 reverse transcriptase increase 3′-azido-3′-deoxythymidine resistance. Proc Natl Acad Sci USA 104(1):317–322
Nowotny M (2009) Retroviral integrase superfamily: the structural perspective. EMBO Rep 10(2):144–151. doi:10.1038/embor.2008.256
Nowotny M, Gaidamakov SA, Crouch RJ, Yang W (2005) Crystal structures of RNase H bound to an RNA/DNA hybrid: substrate specificity and metal-dependent catalysis. Cell 121(7):1005–1016
Nowotny M, Gaidamakov SA, Ghirlando R, Cerritelli SM, Crouch RJ, Yang W (2007) Structure of human RNase H1 complexed with an RNA/DNA hybrid: insight into HIV reverse transcription. Mol Cell 28(2):264–276
Palaniappan C, Fuentes GM, Rodriguez-Rodriguez L, Fay PJ, Bambara RA (1996) Helix structure and ends of RNA/DNA hybrids direct the cleavage specificity of HIV-1 reverse transcriptase RNase H. J Biol Chem 271(4):2063–2070
Parkes KE, Ermert P, Fassler J, Ives J, Martin JA, Merrett JH et al (2003) Use of a pharmacophore model to discover a new class of influenza endonuclease inhibitors. J Med Chem 46(7): 1153–1164
Parniak MA, Min KL, Budihas SR, Le Grice SF, Beutler JA (2003) A fluorescence-based high-throughput screening assay for inhibitors of human immunodeficiency virus-1 reverse transcriptase-associated ribonuclease H activity. Anal Biochem 322(1):33–39. doi:10.1016/j.ab.2003.06.001
Peliska JA, Benkovic SJ (1992) Mechanism of DNA strand transfer reactions catalyzed by HIV-1 reverse transcriptase. [Research Support, U.S. Gov’t, P.H.S.]. Science 258(5085):1112–1118
Rausch JW, Le Grice SF (2004) ‘Binding, bending and bonding’: polypurine tract-primed initiation of plus-strand DNA synthesis in human immunodeficiency virus. Int J Biochem Cell Biol 36(9):1752–1766. doi:10.1016/j.biocel.2004.02.016
Ren J, Nichols C, Bird L, Chamberlain P, Weaver K, Short S et al (2001) Structural mechanisms of drug resistance for mutations at codons 181 and 188 in HIV-1 reverse transcriptase and the improved resilience of second generation non-nucleoside inhibitors. J Mol Biol 312(4):795–805. doi:10.1006/jmbi.2001.4988
Rodgers DW, Gamblin SJ, Harris BA, Ray S, Culp JS, Hellmig B et al (1995) The structure of unliganded reverse transcriptase from the human immunodeficiency virus type 1. Proc Natl Acad Sci USA 92(4):1222–1226
Rothwell PJ, Waksman G (2005) Structure and mechanism of DNA polymerases. Adv Protein Chem 71:401–440. doi:10.1016/S0065-3233(04)71011-6
Ruane PJ, DeJesus E (2004) New nucleoside/nucleotide backbone options: a review of recent studies. J Acquir Immune Defic Syndr 37(Suppl 1):S21–S29
Sarafianos SG, Das K, Tantillo C, Clark AD Jr, Ding J, Whitcomb JM et al (2001) Crystal structure of HIV-1 reverse transcriptase in complex with a polypurine tract RNA:DNA. EMBO J 20(6):1449–1461
Schatz O, Cromme FV, Gruninger-Leitch F, Le Grice SF (1989) Point mutations in conserved amino acid residues within the C-terminal domain of HIV-1 reverse transcriptase specifically repress RNase H function. FEBS Lett 257(2):311–314
Schultz SJ, Champoux JJ (2008) RNase H activity: structure, specificity, and function in reverse transcription. Virus Res 134(1–2):86–103. doi:10.1016/j.virusres.2007.12.007
Schultz SJ, Zhang M, Champoux JJ (2004) Recognition of internal cleavage sites by retroviral RNases H. J Mol Biol 344(3):635–652. doi:10.1016/j.jmb.2004.09.081
Shaw-Reid CA, Munshi V, Graham P, Wolfe A, Witmer M, Danzeisen R et al (2003) Inhibition of HIV-1 ribonuclease H by a novel diketo acid, 4-[5-(benzoylamino)thien-2-yl]-2,4-dioxobutanoic acid. J Biol Chem 278(5):2777–2780. doi:10.1074/jbc.C200621200
Shaw-Reid CA, Feuston B, Munshi V, Getty K, Krueger J, Hazuda DJ et al (2005) Dissecting the effects of DNA polymerase and ribonuclease H inhibitor combinations on HIV-1 reverse-transcriptase activities. Biochemistry 44(5):1595–1606. doi:10.1021/bi0486740
Shih CK, Rose JM, Hansen GL, Wu JC, Bacolla A, Griffin JA (1991) Chimeric human immunodeficiency virus type 1/type 2 reverse transcriptases display reversed sensitivity to nonnucleoside analog inhibitors. Proc Natl Acad Sci USA 88(21):9878–9882
Sluis-Cremer N, Arion D, Parniak MA (2000) Molecular mechanisms of HIV-1 resistance to nucleoside reverse transcriptase inhibitors (NRTIs). Cell Mol Life Sci CMLS 57(10): 1408–1422
Sluis-Cremer N, Arion D, Parniak MA (2002) Destabilization of the HIV-1 reverse transcriptase dimer upon interaction with N-acyl hydrazone inhibitors. Mol Pharmacol 62(2):398–405
Song JJ, Smith SK, Hannon GJ, Joshua-Tor L (2004) Crystal structure of argonaute and its implications for RISC slicer activity. Science 305(5689):1434–1437
Stahl SJ, Kaufman JD, Vikic-Topic S, Crouch RJ, Wingfield PT (1994) Construction of an enzymatically active ribonuclease H domain of human immunodeficiency virus type 1 reverse transcriptase. Protein Eng 7(9):1103–1108
Summa V, Petrocchi A, Pace P, Matassa VG, De Francesco R, Altamura S et al (2004) Discovery of alpha, gamma-diketo acids as potent selective and reversible inhibitors of hepatitis C virus NS5b RNA-dependent RNA polymerase. J Med Chem 47(1):14–17. doi:10.1021/jm0342109
Tan CK, Civil R, Mian AM, So AG, Downey KM (1991) Inhibition of the RNase H activity of HIV reverse transcriptase by azidothymidylate. Biochemistry 30(20):4831–4835
Telesnitsky A, Goff SP (1997) Reverse transcriptase and the generation of retroviral DNA. In: Coffin JM, Hughes SH, Varmus HE (eds) Retroviruses. Cold Spring Harbor Laboratory Press, Plainview, pp. 121–160
Tomassini J, Selnick H, Davies ME, Armstrong ME, Baldwin J, Bourgeois M et al (1994) Inhibition of cap (m7GpppXm)-dependent endonuclease of influenza virus by 4-substituted 2,4-dioxobutanoic acid compounds. Antimicrob Agents Chemother 38(12):2827–2837
Tramontano E, Esposito F, Badas R, Di Santo R, Costi R, La Colla P (2005) 6-[1-(4-Fluorophenyl)methyl-1H-pyrrol-2-yl)]-2,4-dioxo-5-hexenoic acid ethyl ester a novel diketo acid derivative which selectively inhibits the HIV-1 viral replication in cell culture and the ribonuclease H activity in vitro. Antiviral Res 65(2):117–124
Tsunaka Y, Haruki M, Morikawa M, Oobatake M, Kanaya S (2003) Dispensability of glutamic acid 48 and aspartic acid 134 for Mn2+−dependent activity of E. coli ribonuclease HI. Biochemistry 42(11):3366–3374. doi:10.1021/bi0205606
Tsunaka Y, Takano K, Matsumura H, Yamagata Y, Kanaya S (2005) Identification of single Mn(2+) binding sites required for activation of the mutant proteins of E. coli RNase HI at Glu48 and/or Asp134 by X-ray crystallography. J Mol Biol 345(5):1171–1183
Wendeler M, Lee HF, Bermingham A, Miller JT, Chertov O, Bona MK et al (2008) Vinylogous ureas as a novel class of inhibitors of reverse transcriptase-associated ribonuclease H activity. ACS Chem Biol 3(10):635–644
Zhan X, Tan CK, Scott WA, Mian AM, Downey KM, So AG (1994) Catalytically distinct conformations of the ribonuclease H of HIV-1 reverse transcriptase by substrate cleavage patterns and inhibition by azidothymidylate and N-ethylmaleimide. Biochemistry 33(6):1366–1372
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer Science+Business Media New York
About this chapter
Cite this chapter
Rausch, J.W. (2013). Ribonuclease H Inhibitors: Structural and Molecular Biology. In: LeGrice, S., Gotte, M. (eds) Human Immunodeficiency Virus Reverse Transcriptase. Springer, New York, NY. https://doi.org/10.1007/978-1-4614-7291-9_7
Download citation
DOI: https://doi.org/10.1007/978-1-4614-7291-9_7
Published:
Publisher Name: Springer, New York, NY
Print ISBN: 978-1-4614-7290-2
Online ISBN: 978-1-4614-7291-9
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)