Abstract
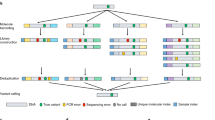
Deep sequencing by NGS of targeted amplicons can identify rare genetic variants in a pool of DNA where the vast majority of genomic DNA does not contain the variant. This approach can be used to detect a previously described paternally inherited, fetal variant in cell-free DNA (cfDNA) in maternal plasma. This is useful in cases where risk for the fetus is contingent upon inheritance of a paternal variant that the woman does not have. Both pathogenic and non-pathogenic variants that the woman does not have can be detected. In cases of compound heterozygosity, presence of the paternal pathogenic variant also requires detection of the maternal variant for risk assessment, which requires a chorion villus biopsy.
We have used this approach to focus on detection of fetal blood groups in cases of presence of maternal alloantibodies against blood group antigens in pregnancy, to predict whether the fetus has inherited a blood group antigen that is targeted by the alloantibodies. In cases of maternal alloantibodies against blood group antigens, the fetus is at risk of hemolytic disease of the fetus and newborn (HDFN). With a known specificity of the maternal antibodies and if the fetal blood group can be determined in the pregnancy, then it can be ascertained if the fetus is at risk of HDFN and rational pregnancy care can be instituted. A noninvasive procedure avoids risks for the fetus. We have reported a procedure based on NGS analysis of PCR amplified cfDNA from maternal plasma. Some fetuses may die as early as week 18. We use this approach to predict fetal K, k, RhC, Rhc, RhE, and ABO blood groups in cases with a risk of HDFN due to the corresponding maternally produced antibodies.
The NGS-based analysis can predict the presence or absence of incompatible antigens on the fetal RBCs.
In this chapter, a noninvasive method for predicting some fetal blood groups early in pregnancy is described. There is a clinical need for such assays, and they may be a useful tool for management of pregnancies complicated by these alloantibodies within the field of precision medicine.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Rieneck K, Bak M, Jønson L, Clausen FB, Krog GR, Tommerup N et al (2013) Next-generation sequencing: proof of concept for antenatal prediction of the fetal K ell blood group phenotype from cell-free fetal DNA in maternal plasma. Transfusion 53(11pt2):2892–2898
Rieneck K, Clausen FB, Bergholt T, Nørgaard LN, Dziegiel MH (2022) Non-invasive fetal K status prediction: 7 years of experience. Transfus Med Hemother 49:1–10
Rieneck K, Clausen FB, Bergholt T, Nørgaard LN, Dziegiel MH (2021) Prenatal prediction of fetal Rh C, c and E status by amplification of maternal cfDNA and deep sequencing. Prenat Diagn 41(11):1380–1388
Rieneck K, Hother CE, Clausen FB, Jakobsen MA, Bergholt T, Hellmuth E et al (2020) Next generation sequencing-based fetal ABO blood group prediction by analysis of cell-free DNA from maternal plasma. Transfus Med Hemother 47(1):45–53
Tsui NB, Jiang P, Chow KC, Su X, Leung TY, Sun H et al (2012) High resolution size analysis of fetal DNA in the urine of pregnant women by paired-end massively parallel sequencing. PLoS One 7(10):e48319. https://doi.org/10.1371/journal.pone.0048319. Epub 20121031. PubMed PMID: 23118982; PubMed Central PMCID: PMCPMC3485143
Rieneck K, Lausen B, Clausen FB, Jønson L, Hansen AT, Dziegiel MH (2022) Exome-based trio analysis for diagnosis of the cause of congenital severe hemolytic anemia in a child. Transfus Med Hemother 49:320–325
Orzinska A, Guz K, Mikula M, Kluska A, Balabas A, Ostrowski J et al (2019) Prediction of fetal blood group and platelet antigens from maternal plasma using next-generation sequencing. Transfusion 59(3):1102–1107. https://doi.org/10.1111/trf.15116. Epub 20190108. PubMed PMID: 30620409
Orzinska A, Kluska A, Balabas A, Piatkowska M, Kulecka M, Ostrowski J et al (2022) Prediction of fetal blood group antigens from maternal plasma using Ion AmpliSeq HD technology. Transfusion 62(2):458–468. https://doi.org/10.1111/trf.16780. Epub 20220108. PubMed PMID: 34997618
O’Brien H, Hyland C, Schoeman E, Flower R, Daly J, Gardener G (2020) Non-invasive prenatal testing (NIPT) for fetal Kell, Duffy and Rh blood group antigen prediction in alloimmunised pregnant women: power of droplet digital PCR. Br J Haematol 189(3):e90–ee4. https://doi.org/10.1111/bjh.16500. Epub 20200216. PubMed PMID: 32062863
Rieneck K, Clausen FB, Dziegiel MH (2015) Noninvasive antenatal determination of fetal blood group using next-generation sequencing. Cold Spring Harb Perspect Med 6(1):a023093. https://doi.org/10.1101/cshperspect.a023093. Epub 20151028. PubMed PMID: 26511760; PubMed Central PMCID: PMCPMC4691807
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Electronic Supplementary Material
Rights and permissions
Copyright information
© 2024 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Rieneck, K. (2024). Cell-Free DNA and Next-Generation Sequencing for Prenatal Diagnosis. In: Félix, L. (eds) Teratogenicity Testing. Methods in Molecular Biology, vol 2753. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-3625-1_38
Download citation
DOI: https://doi.org/10.1007/978-1-0716-3625-1_38
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-3624-4
Online ISBN: 978-1-0716-3625-1
eBook Packages: Springer Protocols