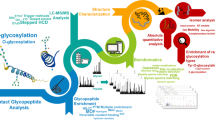
Abstract
Mass spectrometry-driven glycomics and glycoproteomics, the system-wide profiling of detached glycans and intact glycopeptides from biological samples, respectively, are powerful approaches to interrogate the heterogenous glycoproteome. Efforts to develop integrated workflows employing both glycomics and glycoproteomics have been invested since the concerted application of these complementary approaches enables a deeper exploration of the glycoproteome. This protocol paper outlines, step-by-step, an integrated -omics technology, the “glycomics-assisted glycoproteomics” method, that first establishes the N-glycan fine structures and their quantitative distribution pattern of protein extracts via porous graphitized carbon-LC-MS/MS. The N-glycome information is then used to augment and guide the challenging reversed-phase LC-MS/MS-based profiling of intact N-glycopeptides from the same protein samples. Experimental details and considerations relating to the sample preparation and the N-glycomics and N-glycoproteomics data collection, analysis, and integration are discussed. Benefits of the glycomics-assisted glycoproteomics method, which can be readily applied to both simple and complex biological specimens such as protein extracts from cells, tissues, and bodily fluids (e.g., serum), include quantitative information of the protein carriers and site(s) of glycosylation, site occupancy, and the site-specific glycan structures directly from biological samples. The glycomics-assisted glycoproteomics method therefore facilitates a comprehensive view of the complexity and dynamics of the heterogenous glycoproteome.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Varki A (2016) Biological roles of glycans. Glycobiology 27(1):3–49. https://doi.org/10.1093/glycob/cww086
Reily C, Stewart TJ, Renfrow MB, Novak J (2019) Glycosylation in health and disease. Nat Rev Nephrol 15(6):346–366. https://doi.org/10.1038/s41581-019-0129-4
Chernykh A, Kawahara R, Thaysen-Andersen M (2021) Towards structure-focused glycoproteomics. Biochem Soc Trans. https://doi.org/10.1042/BST20200222
Thaysen-Andersen M, Packer NH, Schulz BL (2016) Maturing glycoproteomics technologies provide unique structural insights into the N-glycoproteome and its regulation in health and disease. Mol Cell Proteomics 15(6):1773–1790. https://doi.org/10.1074/mcp.O115.057638
Abrahams JL, Taherzadeh G, Jarvas G, Guttman A, Zhou Y, Campbell MP (2020) Recent advances in glycoinformatic platforms for glycomics and glycoproteomics. Curr Opin Struct Biol 62:56–69. https://doi.org/10.1016/j.sbi.2019.11.009
Hu H, Khatri K, Zaia J (2017) Algorithms and design strategies towards automated glycoproteomics analysis. Mass Spectrom Rev 36(4):475–498. https://doi.org/10.1002/mas.21487
Peng W, Gutierrez Reyes CD, Gautam S, Yu A, Cho BG, Goli M, Donohoo K, Mondello S, Kobeissy F, Mechref Y (2021) MS-based glycomics and glycoproteomics methods enabling isomeric characterization. Mass Spectrom Rev. https://doi.org/10.1002/mas.21713
Gaunitz S, Nagy G, Pohl NL, Novotny MV (2017) Recent advances in the analysis of complex glycoproteins. Anal Chem 89(1):389–413. https://doi.org/10.1021/acs.analchem.6b04343
Thomas DR, Scott NE (2021) Glycoproteomics: growing up fast. Curr Opin Struct Biol 68:18–25. https://doi.org/10.1016/j.sbi.2020.10.028
West CM, Malzl D, Hykollari A, Wilson IBH (2021) Glycomics, glycoproteomics, and glycogenomics: an inter-taxa evolutionary perspective. Mol Cell Proteomics 20:100024. https://doi.org/10.1074/mcp.R120.002263
Thaysen-Andersen M, Kolarich D, Packer NH (2021) Glycomics & Glycoproteomics: from analytics to function. Mol Omics 17(1):8–10. https://doi.org/10.1039/d0mo90019b
Oliveira T, Thaysen-Andersen M, Packer NH, Kolarich D (2021) The Hitchhiker’s guide to glycoproteomics. Biochem Soc Trans 49(4):1643–1662. https://doi.org/10.1042/BST20200879
Bagdonaite I, Malaker SA, Polasky DA, Riley NM, Schjoldager K, Vakhrushev SY, Halim A, Aoki-Kinoshita KF, Nesvizhskii AI, Bertozzi CR, Wandall HH, Parker BL, Thaysen-Andersen M, Scott NE (2022) Glycoproteomics. Nat Rev Methods Primers 2(1):48. https://doi.org/10.1038/s43586-022-00128-4
Rudd PM, Karlsson NG, Khoo KH, Thaysen-Andersen M, Wells L, Packer NH (2022) Glycomics and glycoproteomics. In: Th VA, Cummings RD et al (eds) Essentials of glycobiology, Cold Spring Harbor (NY), pp 689–704. https://doi.org/10.1101/glycobiology.4e.51
Madunić K, Zhang T, Mayboroda OA, Holst S, Stavenhagen K, Jin C, Karlsson NG, Lageveen-Kammeijer GSM, Wuhrer M (2021) Colorectal cancer cell lines show striking diversity of their O-glycome reflecting the cellular differentiation phenotype. Cell Mol Life Sci 78(1):337–350. https://doi.org/10.1007/s00018-020-03504-z
Parker R, Partridge T, Wormald C, Kawahara R, Stalls V, Aggelakopoulou M, Parker J, Powell Doherty R, Ariosa Morejon Y, Lee E, Saunders K, Haynes BF, Acharya P, Thaysen-Andersen M, Borrow P, Ternette N (2021) Mapping the SARS-CoV-2 spike glycoprotein-derived peptidome presented by HLA class II on dendritic cells. Cell Rep 35(8). https://doi.org/10.1016/j.celrep.2021.109179
Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 spike and human ACE2 receptor. Cell Host Microbe 28(4):586–601. e586
Chatterjee S, Kawahara R, Tjondro HC, Shaw DR, Nenke MA, Torpy DJ, Thaysen-Andersen M (2021) Serum N-Glycomics stratifies Bacteremic patients infected with different pathogens. J Clin Med 10(3):516. https://doi.org/10.3390/jcm10030516
Osimanjiang W, Roballo KCS, Houck BD, Ito M, Antonopoulos A, Dell A, Haslam SM, Bushman JS (2020) Analysis of N- and O-linked glycosylation: differential glycosylation after rat spinal cord injury. J Neurotrauma 37(18):1954–1962. https://doi.org/10.1089/neu.2019.6974
Chatterjee S, Ugonotti J, Lee LY, Everest-Dass A, Kawahara R, Thaysen-Andersen M (2021) Trends in oligomannosylation and α1,2-mannosidase expression in human cancers. Oncotarget 12(21):2188–2205. https://doi.org/10.18632/oncotarget.28064
Chang D, Klein JA, Nalehua MR, Hackett WE, Zaia J (2021) Data-independent acquisition mass spectrometry for site-specific glycoproteomics characterization of SARS-CoV-2 spike protein. Anal Bioanal Chem 413(29):7305–7318. https://doi.org/10.1007/s00216-021-03643-7
Pan J, Hu Y, Sun S, Chen L, Schnaubelt M, Clark D, Ao M, Zhang Z, Chan D, Qian J, Zhang H (2020) Glycoproteomics-based signatures for tumor subtyping and clinical outcome prediction of high-grade serous ovarian cancer. Nat Commun 11(1):6139. https://doi.org/10.1038/s41467-020-19976-3
Venkatakrishnan V, Dieckmann R, Loke I, Tjondro HC, Chatterjee S, Bylund J, Thaysen-Andersen M, Karlsson NG, Karlsson-Bengtsson A (2020) Glycan analysis of human neutrophil granules implicates a maturation-dependent glycosylation machinery. J Biol Chem 295(36):12648–12660. https://doi.org/10.1074/jbc.RA120.014011
Sethi MK, Downs M, Shao C, Hackett WE, Phillips JJ, Zaia J (2022) In-depth Matrisome and Glycoproteomic analysis of human brain glioblastoma versus control tissue. Mol Cell Proteomics 21(4):100216. https://doi.org/10.1016/j.mcpro.2022.100216
Saraswat M, Makitie A, Tohmola T, Dickinson A, Saraswat S, Joenvaara S, Renkonen S (2018) Tongue cancer patients can be distinguished from healthy controls by specific N-Glycopeptides found in serum. Proteomics Clin Appl 12(6):e1800061. https://doi.org/10.1002/prca.201800061
Takei D, Harada K, Nouso K, Miyahara K, Dohi C, Matsushita H, Kinugasa H, Hiraoka S, Nishimura SI, Okada H (2022) Clinical utility of a serum glycome analysis in patients with colorectal cancer. J Gastroenterol Hepatol 37(4):727–733. https://doi.org/10.1111/jgh.15781
Reiding KR, Bondt A, Hennig R, Gardner RA, O’Flaherty R, Trbojevic-Akmacic I, Shubhakar A, Hazes JMW, Reichl U, Fernandes DL, Pucic-Bakovic M, Rapp E, Spencer DIR, Dolhain R, Rudd PM, Lauc G, Wuhrer M (2019) High-throughput serum N-Glycomics: method comparison and application to study rheumatoid arthritis and pregnancy-associated changes. Mol Cell Proteomics 18(1):3–15. https://doi.org/10.1074/mcp.RA117.000454
Tomoda T, Nouso K, Kato H, Miyahara K, Dohi C, Morimoto Y, Kinugasa H, Akimoto Y, Matsumoto K, Yamamoto N, Noma Y, Horiguchi S, Tsutsumi K, Amano M, Nishimura S, Yamamoto K (2016) Alteration of serum N-glycan profile in patients with autoimmune pancreatitis. Pancreatology 16(1):44–51. https://doi.org/10.1016/j.pan.2015.11.002
Caval T, Lin YH, Varkila M, Reiding KR, Bonten MJM, Cremer OL, Franc V, Heck AJR (2020) Glycoproteoform profiles of individual patients’ plasma Alpha-1-Antichymotrypsin are unique and extensively remodeled following a septic episode. Front Immunol 11:608466. https://doi.org/10.3389/fimmu.2020.608466
Rosenbalm KE, Tiemeyer M, Wells L, Aoki K, Zhao P (2020) Glycomics-informed glycoproteomic analysis of site-specific glycosylation for SARS-CoV-2 spike protein. STAR Protoc 1(3):100214–100214. https://doi.org/10.1016/j.xpro.2020.100214
Zhao P, Praissman JL, Grant OC, Cai Y, Xiao T, Rosenbalm KE, Aoki K, Kellman BP, Bridger R, Barouch DH, Brindley MA, Lewis NE, Tiemeyer M, Chen B, Woods RJ, Wells L (2020) Virus-receptor interactions of glycosylated SARS-CoV-2 Spike and human ACE2 receptor. Cell Host Microbe 28(4):586–601.e586. https://doi.org/10.1016/j.chom.2020.08.004
Chandler KB, Mehta N, Leon DR, Suscovich TJ, Alter G, Costello CE (2019) Multi-isotype glycoproteomic characterization of serum antibody heavy chains reveals isotype- and subclass-specific N-glycosylation profiles. Mol Cell Proteomics 18(4):686–703. https://doi.org/10.1074/mcp.RA118.001185
Thaysen-Andersen M, Chertova E, Bergamaschi C, Moh ES, Chertov O, Roser J, Sowder R, Bear J, Lifson J, Packer NH, Felber BK, Pavlakis GN (2016) Recombinant human heterodimeric IL-15 complex displays extensive and reproducible N- and O-linked glycosylation. Glycoconj J 33(3):417–433. https://doi.org/10.1007/s10719-015-9627-1
Loke I, Ostergaard O, Heegaard NHH, Packer NH, Thaysen-Andersen M (2017) Paucimannose-rich N-glycosylation of Spatiotemporally regulated human neutrophil elastase modulates its immune functions. Mol Cell Proteomics 16(8):1507–1527. https://doi.org/10.1074/mcp.M116.066746
Loke I, Packer NH, Thaysen-Andersen M (2015) Complementary LC-MS/MS-based N-glycan, N-glycopeptide, and intact N-glycoprotein profiling reveals unconventional Asn71-glycosylation of human neutrophil Cathepsin G. Biomol Ther 5(3):1832–1854. https://doi.org/10.3390/biom5031832
Tjondro HC, Ugonotti J, Kawahara R, Chatterjee S, Loke I, Chen S, Soltermann F, Hinneburg H, Parker BL, Venkatakrishnan V, Dieckmann R, Grant OC, Bylund J, Rodger A, Woods RJ, Karlsson-Bengtsson A, Struwe WB, Thaysen-Andersen M (2021) Hyper-truncated Asn355- and Asn391-glycans modulate the activity of neutrophil granule myeloperoxidase. J Biol Chem 296:100144. https://doi.org/10.1074/jbc.RA120.016342
Parker BL, Thaysen-Andersen M, Solis N, Scott NE, Larsen MR, Graham ME, Packer NH, Cordwell SJ (2013) Site-specific glycan-peptide analysis for determination of N-glycoproteome heterogeneity. J Proteome Res 12(12):5791–5800. https://doi.org/10.1021/pr400783j
Jensen PH, Karlsson NG, Kolarich D, Packer NH (2012) Structural analysis of N- and O-glycans released from glycoproteins. Nat Protoc 7(7):1299–1310. https://doi.org/10.1038/nprot.2012.063
Hinneburg H, Chatterjee S, Schirmeister F, Nguyen-Khuong T, Packer NH, Rapp E, Thaysen-Andersen M (2019) Post-Column Make-Up Flow (PCMF) enhances the performance of capillary-flow PGC-LC-MS/MS-based glycomics. Anal Chem 91(7):4559–4567. https://doi.org/10.1021/acs.analchem.8b05720
Madunic K, Wagt S, Zhang T, Wuhrer M, Lageveen-Kammeijer GSM (2021) Dopant-enriched nitrogen gas for enhanced electrospray ionization of released Glycans in negative ion mode. Anal Chem 93(18):6919–6923. https://doi.org/10.1021/acs.analchem.1c00023
Parker BL, Thaysen-Andersen M, Fazakerley DJ, Holliday M, Packer NH, James DE (2016) Terminal Galactosylation and sialylation switching on membrane glycoproteins upon TNF-alpha-induced insulin resistance in adipocytes. Mol Cell Proteomics 15(1):141–153. https://doi.org/10.1074/mcp.M115.054221
Blazev R, Ashwood C, Abrahams JL, Chung LH, Francis D, Yang P, Watt KI, Qian H, Quaife-Ryan GA, Hudson JE, Gregorevic P, Thaysen-Andersen M, Parker BL (2021) Integrated Glycoproteomics identifies a role of N-glycosylation and Galectin-1 on Myogenesis and muscle development. Mol Cell Proteomics 20:100030. https://doi.org/10.1074/mcp.RA120.002166
Kawahara R, Recuero S, Srougi M, Leite KRM, Thaysen-Andersen M, Palmisano G (2021) The complexity and dynamics of the tissue Glycoproteome associated with prostate cancer progression. Mol Cell Proteomics 20:100026. https://doi.org/10.1074/mcp.RA120.002320
Thaysen-Andersen M, Venkatakrishnan V, Loke I, Laurini C, Diestel S, Parker BL, Packer NH (2015) Human neutrophils secrete bioactive paucimannosidic proteins from azurophilic granules into pathogen-infected sputum. J Biol Chem 290(14):8789–8802. https://doi.org/10.1074/jbc.M114.631622
Kawahara R, Chernykh A, Alagesan K, Bern M, Cao W, Chalkley RJ, Cheng K, Choo MS, Edwards N, Goldman R, Hoffmann M, Hu Y, Huang Y, Kim JY, Kletter D, Liquet B, Liu M, Mechref Y, Meng B, Neelamegham S, Nguyen-Khuong T, Nilsson J, Pap A, Park GW, Parker BL, Pegg CL, Penninger JM, Phung TK, Pioch M, Rapp E, Sakalli E, Sanda M, Schulz BL, Scott NE, Sofronov G, Stadlmann J, Vakhrushev SY, Woo CM, Wu HY, Yang P, Ying W, Zhang H, Zhang Y, Zhao J, Zaia J, Haslam SM, Palmisano G, Yoo JS, Larson G, Khoo KH, Medzihradszky KF, Kolarich D, Packer NH, Thaysen-Andersen M (2021) Community evaluation of glycoproteomics informatics solutions reveals high-performance search strategies for serum glycopeptide analysis. Nat Methods 18(11):1304–1316. https://doi.org/10.1038/s41592-021-01309-x
Palmisano G, Larsen MR, Packer NH, Thaysen-Andersen M (2013) Structural analysis of glycoprotein sialylation – part II: LC-MS based detection. RSC Adv 3(45):22706–22726. https://doi.org/10.1039/C3RA42969E
Young C, Condina MR, Briggs MT, Moh ESX, Kaur G, Oehler MK, Hoffmann P (2021) In-house packed porous graphitic carbon columns for liquid chromatography-mass spectrometry analysis of N-Glycans. Front Chem 9. https://doi.org/10.3389/fchem.2021.653959
Cooper CA, Gasteiger E, Packer NH (2001) GlycoMod--a software tool for determining glycosylation compositions from mass spectrometric data. Proteomics 1(2):340–349. https://doi.org/10.1002/1615-9861(200102)1:23.0.CO;2-B
Ceroni A, Maass K, Geyer H, Geyer R, Dell A, Haslam SM (2008) GlycoWorkbench: a tool for the computer-assisted annotation of mass spectra of glycans. J Proteome Res 7(4):1650–1659. https://doi.org/10.1021/pr7008252
Schilling B, Rardin MJ, MacLean BX, Zawadzka AM, Frewen BE, Cusack MP, Sorensen DJ, Bereman MS, Jing E, Wu CC, Verdin E, Kahn CR, Maccoss MJ, Gibson BW (2012) Platform-independent and label-free quantitation of proteomic data using MS1 extracted ion chromatograms in skyline: application to protein acetylation and phosphorylation. Mol Cell Proteomics 11(5):202–214. https://doi.org/10.1074/mcp.M112.017707
Bern M, Kil YJ, Becker C (2012) Byonic: advanced peptide and protein identification software. Curr Protoc Bioinformatics Chapter 13:Unit13.20. https://doi.org/10.1002/0471250953.bi1320s40
Cao W, Liu M, Kong S, Wu M, Zhang Y, Yang P (2021) Recent advances in software tools for more generic and precise intact glycopeptide analysis. Mol Cell Proteomics 20. https://doi.org/10.1074/mcp.R120.002090
Cox J, Mann M (2008) MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat Biotechnol 26(12):1367–1372. https://doi.org/10.1038/nbt.1511
Cox J, Neuhauser N, Michalski A, Scheltema RA, Olsen JV, Mann M (2011) Andromeda: a peptide search engine integrated into the MaxQuant environment. J Proteome Res 10(4):1794–1805. https://doi.org/10.1021/pr101065j
Saba J, Dutta S, Hemenway E, Viner R (2012) Increasing the productivity of glycopeptides analysis by using higher-energy collision dissociation-accurate mass-product-dependent electron transfer dissociation. Int J Proteomics 2012:560391. https://doi.org/10.1155/2012/560391
Ugonotti J, Kawahara R, Loke I, Zhu Y, Chatterjee S, Tjondro HC, Sumer-Bayraktar Z, Neelamegham S, Thaysen-Andersen M (2021) N-acetyl-β-D-hexosaminidases mediate the generation of paucimannosidic proteins via a putative noncanonical truncation pathway in human neutrophils. Glycobiology 32(3):218–229. https://doi.org/10.1093/glycob/cwab108
Acknowledgments
Prof. Giuseppe Palmisano is thanked for intellectual contributions to the protocol. T.H.C. was supported by an International Macquarie Research Excellence Scholarship (20224231). A.C. was supported by an Australian Government Research Training Program Scholarship. J.U. was supported by a Macquarie University Research Excellence Scholarship. B.L.P. was supported by a University of Melbourne Driving Research Momentum Fellowship. R.K. was supported by a Cancer Institute of New South Wales Fellowship (ECF181259). M.T.-A. was supported by an Australian Research Council Future Fellowship (FT210100455) and a Macquarie University Enterprise Partnership Scheme (175232162).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Chau, T.H., Chernykh, A., Ugonotti, J., Parker, B.L., Kawahara, R., Thaysen-Andersen, M. (2023). Glycomics-Assisted Glycoproteomics Enables Deep and Unbiased N-Glycoproteome Profiling of Complex Biological Specimens. In: Greening, D.W., Simpson, R.J. (eds) Serum/Plasma Proteomics. Methods in Molecular Biology, vol 2628. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2978-9_16
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2978-9_16
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2977-2
Online ISBN: 978-1-0716-2978-9
eBook Packages: Springer Protocols