Abstract
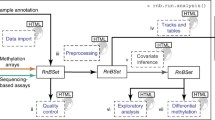
Arrays provide a cost-effective platform for the analysis of human DNA methylation. ShinyÉPICo is an interactive, web-based, and graphical tool that allows the user to analyze Illumina DNA methylation arrays (450 k and EPIC), from the user’s own computer or from a server. This tool covers the analysis entirely, from the raw data input to the final list of differentially methylated positions or regions. Here, we describe the steps of the analysis, the different parameters available, and useful information to understand and select the best options in each step.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Goldberg AD, Allis CD, Bernstein E (2007) Epigenetics: a landscape takes shape. Cell 128:635–638. https://doi.org/10.1016/j.cell.2007.02.006
de la Calle-Fabregat C, Morante-Palacios O, Ballestar E (2020) Understanding the relevance of DNA methylation changes in immune differentiation and disease. Genes (Basel) 11. https://doi.org/10.3390/genes11010110
Jones PA (2012) Functions of DNA methylation: islands, start sites, gene bodies and beyond. Nat Rev Genet 13:484–492. https://doi.org/10.1038/nrg3230
Lev Maor G, Yearim A, Ast G (2015) The alternative role of DNA methylation in splicing regulation. Trends Genet 31:274–280
Chow JC, Yen Z, Ziesche SM, Brown CJ (2005) Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet 6:69–92
Lister R, Pelizzola M, Dowen RH et al (2009) Human DNA methylomes at base resolution show widespread epigenomic differences. Nature 462:315–322. https://doi.org/10.1038/nature08514
Neri F, Rapelli S, Krepelova A et al (2017) Intragenic DNA methylation prevents spurious transcription initiation. Nature 543:72–77. https://doi.org/10.1038/nature21373
Smith ML, Baggerly KA, Bengtsson H et al (2013) Illuminaio: an open source IDAT parsing tool for Illumina microarrays. F1000Research 2:264. https://doi.org/10.12688/f1000research.2-264.v1
Aryee MJ, Jaffe AE, Corrada-Bravo H et al (2014) Minfi: a flexible and comprehensive Bioconductor package for the analysis of Infinium DNA methylation microarrays. Bioinformatics 30:1363–1369. https://doi.org/10.1093/bioinformatics/btu049
Müller F, Scherer M, Assenov Y et al (2019) RnBeads 2.0: comprehensive analysis of DNA methylation data. Genome Biol 20:55. https://doi.org/10.1186/s13059-019-1664-9
Ritchie ME, Phipson B, Wu D et al (2015) Limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43:e47. https://doi.org/10.1093/nar/gkv007
Du P, Kibbe WA, Lin SM (2008) lumi: a pipeline for processing Illumina microarray. Bioinformatics 24:1547–1548. https://doi.org/10.1093/bioinformatics/btn224
Morante-Palacios O, Ballestar E (2021) shinyÉPICo: a graphical pipeline to analyze Illumina DNA methylation arrays. Bioinformatics. https://doi.org/10.1093/bioinformatics/btaa1095
Ritchie ME, Diyagama D, Neilson J et al (2006) Empirical array quality weights in the analysis of microarray data. BMC Bioinf 7:261. https://doi.org/10.1186/1471-2105-7-261
Law CW, Chen Y, Shi W, Smyth GK (2014) Voom: precision weights unlock linear model analysis tools for RNA-seq read counts. Genome Biol 15:R29. https://doi.org/10.1186/gb-2014-15-2-r29
Phipson B, Lee S, Majewski IJ et al (2016) Robust hyperparameter estimation protects against hypervariable genes and improves power to detect differential expression. Ann Appl Stat 10:946–963. https://doi.org/10.1214/16-AOAS920
Martorell-Marugán J, González-Rumayor V, Carmona-Sáez P (2019) MCSEA: detecting subtle differentially methylated regions. Bioinformatics 35:3257–3262. https://doi.org/10.1093/bioinformatics/btz096
McLean CY, Bristor D, Hiller M et al (2010) GREAT improves functional interpretation of cis-regulatory regions. Nat Biotechnol 28:495–501. https://doi.org/10.1038/nbt.1630
Heinz S, Benner C, Spann N et al (2010) Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell 38:576–589. https://doi.org/10.1016/j.molcel.2010.05.004
Funding
O.M.-P. holds an i-PFIS PhD Fellowship [IFI17/00034] from Acción Estratégica en Salud 2013–2016 ISCIII, co-financed by Fondo Social Europeo.
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2023 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Morante-Palacios, O. (2023). Interactive DNA Methylation Array Analysis with ShinyÉPICo. In: Oliveira, P.H. (eds) Computational Epigenomics and Epitranscriptomics. Methods in Molecular Biology, vol 2624. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2962-8_2
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2962-8_2
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2961-1
Online ISBN: 978-1-0716-2962-8
eBook Packages: Springer Protocols