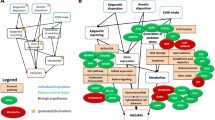
Abstract
Fetal alcohol spectrum disorder (FASD) is a clinically diverse teratogenic disorder with many underlying contributing risk factors including other environmental influences and temporal, epigenetic, and genetic factors. Twin studies of children with FASD have established a genetic component to the disorder; however, identifying which genes and their variants contribute to the pathogenesis of this complex and common disorder remains elusive. In this chapter, we highlight the genetic determinants of rare neurodevelopmental disorders that share overlapping clinical phenotypes to FASD, including CHARGE, Aarskog, Smith-Lemli-Optiz, Opitz-Kaveggia, Campomelic dysplasia, Noonan, Cornelia de Lange, and 22q11.2 deletion syndrome (DS). We hypothesize that by examining the genetic determinants of these rare developmental disorders, we can identify potential prenatal alcohol exposure susceptibility genes of FASD. Indeed, our investigation identified many specific candidate genes in neurodevelopmental signaling pathways that are well established in animal models of FASD-like outcomes following prenatal alcohol exposure, including those in the RAS/MAPK, Wnt/Ca2+, SHH, cholesterol, and retinoic acid pathways. Our findings support the notion that causative alleles of these rare developmental disorders may be sensitizing to prenatal alcohol exposure outcomes as seen in FASD. Sensitization by prenatal alcohol exposure could result in a direct functional modulation of protein activity or induction of an epigenetic modulation of candidate gene expression and define new underlying etiologies of FASD. Moreover, the frequency of PAE-sensitizing variants should be enriched in children with FASD and may provide new FASD diagnostic biomarkers.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
Abbreviations
- 22q11.2DS :
-
22q11.2 Deletion syndrome
- 7-DHC:
-
7-dehydrocholestrol
- BMP:
-
Bone morphogenetic protein
- Ca2+:
-
Calcium
- CamK:
-
Calcium and calmodulin-dependent kinase
- CAV:
-
Caveolin
- CdLS :
-
Cornelia de Lange syndrome
- FASD :
-
Fetal alcohol spectrum disorder
- FGF:
-
Fibroblast growth factor
- LCR:
-
Low-copy number repeats
- MED :
-
Mammalian mediator complex
- NCC :
-
Neural crest cells
- NS :
-
Noonan syndrome
- PAE :
-
Prenatal alcohol exposure
- PTCH:
-
Patched
- RA :
-
Retinoic acid
- RAR:
-
Retinoic acid receptor
- SHH :
-
Sonic hedgehog
- SLOS :
-
Smith-Lemli-Opitz syndrome
- SMO:
-
Smoothened
- TGF-β:
-
Transforming growth factor β
- WNT :
-
Wingless/integrated
References
May PA, Chambers CD, Kalberg WO et al (2018) Prevalence of fetal alcohol spectrum disorders in 4 US communities. JAMA 319:474–482. https://doi.org/10.1001/jama.2017.21896
Zhang X, Sliwowska JH, Weinberg J (2005) Prenatal alcohol exposure and fetal programming: effects on neuroendocrine and immune function. Exp Biol Med 230:376–388. https://doi.org/10.1177/15353702-0323006-05
Mattson S et al (2011) Fetal alcohol spectrum disorders: neuropsychological and behavioral features. Neuropsychol Rev 21:81–101. https://doi.org/10.1007/s11065-011-9167-9.Fetal
Pei J, Denys K, Hughes J, Rasmussen C (2011) Mental health issues in fetal alcohol spectrum disorder. J Ment Health 20:473–483. https://doi.org/10.3109/09638237.2011.577113
Jones KL, Smith DW (1973) Recognition of the fetal alcohol syndrome in early infancy. Lancet 302:999–1001. https://doi.org/10.1016/S0140-6736(73)91092-1
Astley SJ (2000) Diagnosing the full Spectrum of fetal alcohol-exposed individuals: introducing the 4-digit diagnostic code. Alcohol Alcohol 35:400–410. https://doi.org/10.1093/alcalc/35.4.400
DeMyer W, Zeman WPC (1964) The face predicts the brain: diagnostic significance of median facial anomalies for holoprosencephaly (arhinencephaly). Pediatrics 34:256–263
Eberhart J, Parnell S (2016) The genetics of fetal alcohol spectrum disorders (FASD). Alcohol Clin Exp Res 40:1154–1165. https://doi.org/10.1111/acer.13066.The
Streissguth AP, Dehaene P (1993) Fetal alcohol syndrome in twins of alcoholic mothers: concordance of diagnosis and IQ. Am J Med Genet 47:857–861. https://doi.org/10.1002/ajmg.1320470612
Chen Y, Ozturk NC, Ni L et al (2011) Strain differences in developmental vulnerability to alcohol exposure via embryo culture in mice. Alcohol Clin Exp Res 35:1293–1304. https://doi.org/10.1111/j.1530-0277.2011.01465.x
Debelak KA, Smith SM (2000) Avian genetic background modulates the neural crest apoptosis induced by ethanol exposure. Alcohol Clin Exp Res 24:307–314. https://doi.org/10.1097/00000374-200003000-00008
Loucks E, Carvan MJ (2004) Strain-dependent effects of developmental ethanol exposure in zebrafish. Neurotoxicol Teratol 26:745–755. https://doi.org/10.1016/j.ntt.2004.06.017
McDonald-McGinn DM, Sullivan KE, Marino B et al (2015) 22Q11.2 deletion syndrome. Nat Rev Dis Prim 1. https://doi.org/10.1038/nrdp.2015.71
Hemingway SJA, Bledsoe JM, Davies JK et al (2019) Twin study confirms virtually identical prenatal alcohol exposures can lead to markedly different fetal alcohol spectrum disorder outcomes- fetal genetics influences fetal vulnerability. Adv Pediatr Res 5:1–19. https://doi.org/10.24105/apr.201
Yelin R, Schyr RBH, Kot H et al (2005) Ethanol exposure affects gene expression in the embryonic organizer and reduces retinoic acid levels. Dev Biol 279:193–204. https://doi.org/10.1016/j.ydbio.2004.12.014
Yelin R, Kot H, Yelin D, Fainsod A (2007) Early molecular effects of ethanol during vertebrate embryogenesis. Differentiation 75:393–403. https://doi.org/10.1111/j.1432-0436.2006.00147.x
Green ML, Singh AV, Yihzi Z et al (2007) Reprogramming of genetic networks during initiation of the Fetal Alcohol Syndrome. Dev Dyn 236:613–631
Hong M, Krauss RS (2012) Cdon mutation and fetal ethanol exposure synergize to produce midline signaling defects and holoprosencephaly spectrum disorders in mice. PLoS Genet 8. https://doi.org/10.1371/journal.pgen.1002999
Garic A, Berres ME, Smith SM (2014) High-throughput transcriptome sequencing identifies candidate genetic modifiers of vulnerability to fetal alcohol spectrum disorders. Alcohol Clin Exp Res 38:1874–1882. https://doi.org/10.1111/acer.12457
Kietzman HW, Everson JL, Sulik KK, Lipinski RJ (2014) The teratogenic effects of prenatal ethanol exposure are exacerbated by sonic hedgehog or Gli2 haploinsufficiency in the mouse the teratogenic effects of prenatal ethanol exposure are exacerbated by sonic hedgehog or Gli2 haploinsufficiency in the mouse. PLoS One. https://doi.org/10.1371/journal.pone.0089448
Smith SM, Garic A, Flentke GR, Berres ME (2014) Neural crest development in fetal alcohol syndrome. Birth Defects Res C Embryo Today 102:210–220. https://doi.org/10.1002/bdrc.21078
Sarmah S, Muralidharan P, Marrs JA (2016) Embryonic ethanol exposure dysregulates bmp and notch signaling, leading to persistent atrio-ventricular valve defects in zebrafish. PLoS One 11:1–28. https://doi.org/10.1371/journal.pone.0161205
Petrelli B, Weinberg J, Hicks GG (2018) Effects of prenatal alcohol exposure (PAE): insights into FASD using mouse models of PAE. Biochem Cell Biol 96:131–147. https://doi.org/10.1139/bcb-2017-0280
Ahlgren SC, Thakur V, Bronner-Fraser M (2002) Sonic hedgehog rescues cranial neural crest from cell death induced by ethanol exposure. Proc Natl Acad Sci U S A 99:10476–10481. https://doi.org/10.1073/pnas.162356199
Li Y, Yang H, Zdanowicz M et al (2007) Fetal alcohol exposure impairs hedgehog cholesterol modification and signaling. Lab Investig 87:231–240. https://doi.org/10.1038/labinvest.3700516
Aoto K, Shikata Y, Higashiyama D et al (2008) Fetal ethanol exposure activates protein kinase a and impairs Shh expression in prechordal mesendoderm cells in the pathogenesis of holoprosencephaly. Birth Defects Res A Clin Mol Teratol 82:224–231. https://doi.org/10.1002/bdra.20447
Serrano M, Han M, Brinez P, Linask KK (2010) Fetal alcohol syndrome: cardiac birth defects in mice and prevention with folate. Am J Obstet Gynecol 203:75.e7–75.e15. https://doi.org/10.1016/j.ajog.2010.03.017
Muralidharan P, Sarmah S, Marrs JA (2015) Zebrafish retinal defects induced by ethanol exposure are rescued by retinoic acid and folic acid supplement. Alcohol 49:149–163. https://doi.org/10.1016/j.alcohol.2014.11.001
Zhang P, Wang G, Lin Z et al (2017) Alcohol exposure induces chick craniofacial bone defects by negatively affecting cranial neural crest development. Toxicol Lett 281:53–64. https://doi.org/10.1016/j.toxlet.2017.09.010
Shabtai Y, Fainsod A (2018) Competition between ethanol clearance and retinoic acid biosynthesis in the induction of fetal alcohol syndrome. Biochem Cell Biol 96:148–160. https://doi.org/10.1139/bcb-2017-0132
Chiang C, Litingtung Y, Lee E et al (1996) Cyclopia and defective axial patterning in mice lacking Sonic hedgehog gene function. Nature 383:407–413
Ming JE, Roessler E, Muenke M (1998) Human developmental disorders and the Sonic hedgehog pathway. Mol Med Today 4310:343–349
Yamagishi C, Yamagishi H, Maeda J et al (2006) Sonic hedgehog is essential for first pharyngeal arch development. Pediatr Res 59:349–354. https://doi.org/10.1203/01.pdr.0000199911.17287.3e
Heyne GW, Melberg CG, Doroodchi P et al (2015) Definition of critical periods for hedgehog pathway antagonist-induced holoprosencephaly, cleft lip, and cleft palate. PLoS One 10:1–14, e0120517. https://doi.org/10.1371/journal.pone.0120517
Dworkin S, Boglev Y, Owens H, Goldie SJ (2016) The role of sonic hedgehog in craniofacial patterning, morphogenesis and cranial neural crest survival. J Dev Biol 4:24. https://doi.org/10.3390/jdb4030024
Yamada Y, Nagase T, Nagase M, Koshima I (2005) Gene expression changes of sonic hedgehog signaling cascade in a mouse embryonic model of fetal alcohol syndrome. J Craniofac Surg 16:1055–1061
Gurdon JB, Bourillot P-Y (2001) Morphogen gradient interpretation. Nature 413:797–803
Logan CY, Nusse R (2004) The Wnt signaling pathway in development and disease. Annu Rev Cell Dev Biol 20:781–810. https://doi.org/10.1146/annurev.cellbio.20.010403.113126
Arenzana FJ, Iii MJC, Aijón J et al (2006) Teratogenic effects of ethanol exposure on zebrafish visual system development. Neurotoxicol Teratol 28:342–348. https://doi.org/10.1016/j.ntt.2006.02.001
Muralidharan P, Sarmah S, Zhou FC, Marrs JA (2013) Fetal alcohol spectrum disorder (FASD) associated neural defects: complex mechanisms and potential therapeutic targets. Brain Sci 3:964–991. https://doi.org/10.3390/brainsci3020964
Sarmah S, Marrs JA (2013) Complex cardiac defects after ethanol exposure during discrete cardiogenic events in zebrafish: prevention with folic acid. Dev Dyn 242:1184–1201. https://doi.org/10.1002/dvdy.24015
Swartz ME, Wells MB, Griffin M et al (2014) A screen of zebrafish mutants identifies ethanol-sensitive genetic loci. Alcohol Clin Exp Res 38:694–703. https://doi.org/10.1111/acer.12286
Ben LC, Fernandes Y, Eberhart JK (2016) Fishing for fetal alcohol spectrum disorders: zebrafish as a model for ethanol teratogenesis. Zebrafish 13:391–398. https://doi.org/10.1089/zeb.2016.1270
Lauing KL, Roper PM, Nauer RK, Callaci JJ (2012) Acute alcohol exposure impairs fracture healing and deregulates β-catenin signaling in the fracture callus. Alcohol Clin Exp Res 36:2095–2103. https://doi.org/10.1111/j.1530-0277.2012.01830.x
Garic A, Flentke GR, Amberger E et al (2011) CaMKII activation is a novel effector of alcohol’s neurotoxicity in neural crest stem/progenitor cells. J Neurochem 118:646–657. https://doi.org/10.1111/j.1471-4159.2011.07273.x
Reynolds K, Kumari P, Rincon LS et al (2019) Wnt signaling in orofacial clefts: crosstalk, pathogenesis and models. Dis Model Mech 12:dmm037051. https://doi.org/10.1242/dmm.037051
Johnson CS, Zucker RM, Sidney E et al (2007) Perturbation of retinoic acid (RA) –mediated limb development suggests a role for diminished RA signaling in the teratogenesis of ethanol. Birth Defects Res (Part A) 641:631–641. https://doi.org/10.1002/bdra.20385
Marrs JA, Clendenon SG, Ratcliffe DR et al (2010) Zebrafish fetal alcohol syndrome model: effects of ethanol are rescued by retinoic acid supplement. Alcohol 44:707–715. https://doi.org/10.1016/j.alcohol.2009.03.004
Shabtai Y, Bendelac L, Jubran H et al (2018) Acetaldehyde inhibits retinoic acid biosynthesis to mediate alcohol teratogenicity. Sci Rep 8:1–14. https://doi.org/10.1038/s41598-017-18719-7
Xu Q, Kopp JB (2012) Retinoid and TGF-β families: crosstalk in development, neoplasia, immunity, and tissue repair. Semin Nephrol 32:287–294. https://doi.org/10.1016/j.semnephrol.2012.04.008
Malgorzata S, Nowaczyk JM (2020) Smith-Lemli-Opitz syndrome summary. NIH Gene Rev 1–24
Zhou C, Chen J, Zhang X et al (2014) Prenatal ethanol exposure up-regulates the cholesterol transporters ATP-binding cassette A1 and G1 and reduces cholesterol levels in the developing rat. Brain 49:626–634. https://doi.org/10.1093/alcalc/agu049
Jamuar SS, Picker JD, Stoler JM (2018) Utility of genetic testing in fetal alcohol spectrum disorder. J Pediatr 196:270–274.e1. https://doi.org/10.1016/j.jpeds.2017.12.046
Douzgou S, Breen C, Crow YJ et al (2012) Diagnosing fetal alcohol syndrome: new insights from newer genetic technologies. Arch Dis Child 97:812–817. https://doi.org/10.1136/archdischild-2012-302125
Abdelmalik N, Van Haelst M, Mancini G et al (2013) Diagnostic outcomes of 27 children referred by pediatricians to a genetics clinic in the Netherlands with suspicion of fetal alcohol spectrum disorders. Am J Med Genet Part A 161A:254–260. https://doi.org/10.1002/ajmg.a.35672
Qin Z, Su J, Li M et al (2020) Clinical and genetic analysis of CHD7 expands the genotype and phenotype of CHARGE syndrome. Front Genet 11:1–5. https://doi.org/10.3389/fgene.2020.00592
Zentner GE, Layman WS, Martin DM, Scacheri PC (2010) Molecular and phenotypic aspects of CHD7 mutation in CHARGE syndrome. Am J Med Genet A 152(A):674–686. https://doi.org/10.1002/ajmg.a.33323.Molecular
Bouazoune K, Kingston RE (2012) Chromatin remodeling by the CHD7 protein is impaired by mutations that cause human developmental disorders. PNAS 109:1–6. https://doi.org/10.1073/pnas.1213825109. www.pnas.org/cgi/doi/10.1073/pnas.1213825109
Sarmah S, Srivastava R, Mcclintick JN et al (2020) Embryonic ethanol exposure alters expression of sox2 and other early transcripts in zebrafish, producing gastrulation defects. Sci Rep 10:1–13. https://doi.org/10.1038/s41598-020-59043-x
Engelen E, Akinci U, Bryne JC et al (2011) Sox2 cooperates with Chd7 to regulate genes that are mutated in human syndromes. Nat Genet 43:607–612. https://doi.org/10.1038/ng.825
Chrisman K, Kenney R, Comin J et al (2004) Gestational ethanol exposure disrupts the expression of FGF8 and sonic hedgehog during limb patterning. Birth Defects Res (Part A) Clin Mol Teratol 171:163–171. https://doi.org/10.1002/bdra.20019
Yu T, Meiners LC, Danielsen K, et al (2013) Deregulated FGF and homeotic gene expression underlies cerebellar vermis hypoplasia in CHARGE syndrome. elife 2. https://doi.org/10.7554/eLife.01305
Kot-Leibovich H, Fainsod A (2009) Ethanol induces embryonic malformations by competing for retinaldehyde dehydrogenase activity during vertebrate gastrulation. Dis Model Mech 2:295–305. https://doi.org/10.1242/dmm.001420
Yao H, Hill SF, Skidmore JM et al (2018) CHD7 represses the retinoic acid synthesis enzyme ALDH1A3 during inner ear development. JCI Insight 3. https://doi.org/10.1172/JCI.INSIGHT.97440
Balendran V, Skidmore JM, Ritter KE et al (2021) Chromatin remodeler CHD7 is critical for cochlear morphogenesis and neurosensory patterning. Dev Biol 477:11–21. https://doi.org/10.1016/j.ydbio.2021.05.009
Higashida H, Yokoyama S, Huang J et al (2012) Neurochemistry international social memory, amnesia, and autism: brain oxytocin secretion is regulated by NAD + metabolites and single nucleotide polymorphisms of CD38. Neurochem Int 61:828–838. https://doi.org/10.1016/j.neuint.2012.01.030
Fujita K, Ogawa R, Kawawaki S, Ito K (2014) Roles of chromatin remodelers in maintenance mechanisms of multipotency of mouse trunk neural crest cells in the formation of neural crest-derived stem cells. Mech Dev 133:126–145. https://doi.org/10.1016/j.mod.2014.05.001
Bajpai R, Chen DA, Rada-iglesias A et al (2010) CHD7 cooperates with PBAF to control multipotent neural crest formation. Nature 463:3–9. https://doi.org/10.1038/nature08733
Jiang X, Zhou Y, Xian L, Chen W (2012) The mutation in Chd7 causes misexpression of Bmp4 and developmental defects in telencephalic midline. Am J Pathol 181:626–641. https://doi.org/10.1016/j.ajpath.2012.05.006
Feng W, Kawauchi D, Ko H et al (2017) Chd7 is indispensable for mammalian brain development through activation of a neuronal differentiation programme. Nat Commun 8. https://doi.org/10.1038/ncomms14758
Parnell SE, O’Leary-Moore SK, Godin EA, Dehart DB, Johnson BW, Johnson GA, Styner MA, Sulik KK (2009) Magnetic resonance microscopy defines ethanol-induced brain abnormalities in prenatal mice: effects of acute insult on gestational day 8. Alcohol Clin Exp Res 33:1001–1011
Godin EA, O’Leary-Moore SK, Khan AA et al (2010) Magnetic resonance microscopy defines ethanol-induced brain abnormalities in prenatal mice: effects of acute insult on gestational day 7. Alcohol Clin Exp Res. https://doi.org/10.1111/j.1530-0277.2009.01071.x
Gage PJ, Hurd EA, Martin DM (2015) Mouse models for the dissection of CHD7 functions in eye development and the molecular basis for ocular defects in CHARGE syndrome. IOVS 56:7923–7930. https://doi.org/10.1167/iovs.15-18069
Dunty WC Jr, Zucker RM, Sulik KK (2002) Hindbrain and cranial nerve dysmorphogenesis result from acute maternal ethanol administration. Dev Neurosci 24:328–342
Randall V, Mccue K, Roberts C et al (2009) Great vessel development requires biallelic expression of Chd7 and Tbx1 in pharyngeal ectoderm in mice. J Clin Invest 119:3301–3310. https://doi.org/10.1172/JCI37561.some
Asad Z, Pandey A, Babu A et al (2016) Rescue of neural crest-derived phenotypes in a zebrafish CHARGE model by Sox10 downregulation. Hum Mol Genet 25:3539–3554. https://doi.org/10.1093/hmg/ddw198
Dunty WC, Zucker RM, Sulik K (2002) Hindbrain and cranial nerve dysmorphogenesis result from acute maternal ethanol administration. Dev Neurosci 7090:328–342. https://doi.org/10.1159/000066748
Orrico A, Galli L, Obregon MG et al (2007) Unusually severe expression of craniofacial features in Aarskog-Scott syndrome due to a novel truncating mutation of the FGD1 gene. Am J Med Genet Part A 143A:58–63. https://doi.org/10.1002/ajmg.a.31562
Orrico A, Galli L, Clayton-Smith J, Fryns JP (2015) Clinical utility gene card for: Aarskog-Scott Syndrome (faciogenital dysplasia) – update 2015. Eur J Hum Genet 23:10–13. https://doi.org/10.1038/ejhg.2014.178
Orrico A, Galli L, Cavaliere ML et al (2004) Phenotypic and molecular characterisation of the Aarskog-Scott syndrome: a survey of the clinical variability in light of FGD1 mutation analysis in 46 patients. Eur J Hum Genet 12:16–23. https://doi.org/10.1038/sj.ejhg.5201081
Gorski JL, Estrada L, Hu C, Liu Z (2000) Skeletal-specific expression of Fgd1 during bone formation and skeletal defects in faciogenital dysplasia (FGDY; Aarskog syndrome). Dev Dyn 586:573–586
Estrada L, Caron E, Gorski JL (2001) Fgd1, the Cdc42 guanine nucleotide exchange factor responsible for faciogenital dysplasia, is localized to the subcortical actin cytoskeleton and Golgi membrane. Hum Mol Genet 10:485–496
Whitehead IP, Abe K, Gorski JL, Der CJ (1998) CDC42 and FGD1 cause distinct signaling and transforming activities. Mol Cell Biol 18:4689–4697. https://doi.org/10.1128/mcb.18.8.4689
Zheng Y, Fischer DJ, Santos MF et al (1996) The faciogenital dysplasia gene product FGD1 functions as a Cdc42Hs-specific guanine-nucleotide exchange factor. J Biol Chem 271:33169–33172. https://doi.org/10.1074/jbc.271.52.33169
Liu Y, Jin Y, Li J et al (2013) Inactivation of Cdc42 in neural crest cells causes craniofacial and cardiovascular morphogenesis defects. Dev Biol 383:239–252. https://doi.org/10.1016/j.ydbio.2013.09.013
Carey MB, Matsumoto SG (1999) Spontaneous calcium transients are required for neuronal differentiation of murine neural crest. Dev Biol 215:298–313. https://doi.org/10.1006/dbio.1999.9433
Garic-stankovic A, Hernandez MR, Chiang PJ et al (2005) Ethanol triggers neural crest apoptosis through the selective activation of a pertussis toxin – sensitive G protein and a phospholipase Cbeta-dependent Ca2+ transient. Alcohol Clin Exp Res 29:1237–1246. https://doi.org/10.1097/01.ALC.0000172460.05756.D9
Carey MB, Matsumoto SG (2000) Calcium transient activity in cultured murine neural crest cells is regulated at the IP3 receptor. Brain Res 862:201–210. https://doi.org/10.1016/S0006-8993(00)02128-4
Fitzky BU, Witsch-Baumgartner M, Erdel M et al (1998) Mutations in the Δ7-sterol reductase gene in patients with the Smith-Lemli-Opitz syndrome. Proc Natl Acad Sci U S A 95:8181–8186. https://doi.org/10.1073/pnas.95.14.8181
Nowaczyk MJM, Tan M, Hamid JS, Allanson JE (2012) Smith-Lemli-Opitz syndrome: objective assessment of facial phenotype. Am J Med Genet Part A 158(A):1020–1028. https://doi.org/10.1002/ajmg.a.35285
Kline AD, Grados M, Sponseller P, Levy HP (2007) Natural history of aging in Cornelia de Lange syndrome. Am J Med Genet C Semin Med Genet 145C:248–260. https://doi.org/10.1002/ajmg.c.30137.Natural
Yu H, Patel SB (2005) Recent insights into the Smith-Lemli-Opitz syndrome. Clin Genet 68:383–391. https://doi.org/10.1111/j.1399-0004.2005.00515.x
Battaile KP, Battaile BC, Merkens LS et al (2001) Carrier frequency of the common mutation IVS8-1G>C in DHCR7 and estimate of the expected incidence of Smith–Lemli–Opitz syndrome. Mol Genet Metab 72:67–71. https://doi.org/10.1006/mgme.2000.3103
Bailey BA, Sokol RJ (2011) Prenatal alcohol exposure and miscarriage, stillbirth, preterm delivery, and sudden infant death syndrome. Alcohol Res Health 34:86–91
Prabhu AV, Luu W, Sharpe LJ, Brown AJ (2016) Cholesterol-mediated degradation of 7-dehydrocholesterol reductase switches the balance from cholesterol to Vitamin D synthesis. J Biol Chem 291:8363–8376. https://doi.org/10.1074/jbc.M115.699546
Zuin J, Franke V, van Ijcken WFJ et al (2014) A Cohesin-independent role for NIPBL at promoters provides insights in CdLS. PLoS Genet 10. https://doi.org/10.1371/journal.pgen.1004153
Suzuki A, Ogata K, Yoshioka H et al (2020) Disruption of Dhcr7 and Insig1/2 in cholesterol metabolism causes defects in bone formation and homeostasis through primary cilium formation. Bone Res 8. https://doi.org/10.1038/s41413-019-0078-3
Yusifov E, Dumoulin A, Stoeckli ET (2021) Investigating primary cilia during peripheral nervous system formation. Int J Mol Sci 22:1–20. https://doi.org/10.3390/ijms22063176
Emmer BT, Maric D, Engman DM (2010) Molecular mechanisms of protein and lipid targeting to ciliary membranes. J Cell Sci 123:529–536. https://doi.org/10.1242/jcs.062968
Radhakrishnan A, Rohatgi R, Siebold C (2020) Cholesterol access in cellular membranes controls Hedgehog signaling. Nat Chem Biol 16:1303–1313. https://doi.org/10.1038/s41589-020-00678-2
Goetz SC, Ocbina PJR, Anderson KV (2009) The primary cilium as a hedgehog signal transduction machine. Methods Cell Biol 94:199–222. https://doi.org/10.1016/S0091-679X(08)94010-3
Lipinski RJ, Godin EA, Leary-moore SKO et al (2010) Genesis of teratogen-induced holoprosencephaly in mice. Am J Med Genet Part C 42:29–42. https://doi.org/10.1002/ajmg.c.30239
Smith TG, Laval S, Chen F et al (2014) Neural crest cell-specific inactivation of Nipbl or Mau2 during mouse development results in a late onset of craniofacial defects. Genesis 52:687–694. https://doi.org/10.1002/dvg.22780
Rubinato E, Rondeau S, Giuliano F et al (2020) MED12 missense mutation in a three-generation family. Clinical characterization of MED12-related disorders and literature review. Eur J Med Genet Genet 63:103768. https://doi.org/10.1016/j.ejmg.2019.103768
Knuesel MT, Meyer KD, Bernecky C, Taatjes DJ (2009) The human CDK8 subcomplex is a molecular switch that controls mediator coactivator function. Genes Dev 23:439–451. https://doi.org/10.1101/gad.1767009
Ding N, Zhou H, Esteve PO et al (2008) Mediator links epigenetic silencing of neuronal gene expression with X-linked mental retardation. Mol Cell 31:347–359. https://doi.org/10.1016/j.molcel.2008.05.023
Donnio L, Bidon B, Hashimoto S et al (2017) MED12- related XLID disorders are dose-dependent of immediate early genes (IEGs) expression. Hum Mol Genet 26:2062–2075. https://doi.org/10.1093/hmg/ddx099
Wang H, Shen Q, Ye LH, Ye J (2013) MED12 mutations in human diseases. Protein Cell 4:643–646. https://doi.org/10.1007/s13238-013-3048-3
Mo X, Kowenz-Leutz E, Xu H, Leutz A (2004) Ras induces mediator complex exchange on C/EBPβ. Mol Cell 13:241–250. https://doi.org/10.1016/S1097-2765(03)00521-5
Pavri R, Lewis B, Kim TK et al (2005) PARP-1 determines specificity in a retinoid signaling pathway via direct modulation of mediator. Mol Cell 18:83–96. https://doi.org/10.1016/j.molcel.2005.02.034
Csukasi F, Duran I, Zhang W et al (2019) Dominant-negative SOX9 mutations in campomelic dysplasia. Hum Mutat 40:2344–2352. https://doi.org/10.1002/humu.23888
Castori M, Bottillo I, Morlino S et al (2016) Variability in a three-generation family with Pierre Robin sequence, acampomelic campomelic dysplasia, and intellectual disability due to a novel ∼1 Mb deletion upstream of SOX9, and including KCNJ2 and KCNJ16. Birth Defects Res Part A Clin Mol Teratol 106:61–68. https://doi.org/10.1002/bdra.23463
Jo A, Denduluri S, Zhang B et al (2014) The versatile functions of Sox9 in development, stem cells, and human diseases. Genes Dis 1:149–161. https://doi.org/10.1016/j.gendis.2014.09.004
Thouvenin B, Djadi-Prat J, Chalouhi C et al (2013) Developmental outcome in Pierre Robin sequence: a longitudinal and prospective study of a consecutive series of severe phenotypes. Am J Med Genet Part A 161:312–319. https://doi.org/10.1002/ajmg.a.35773
Evans KN, Sie KC, Hopper RA et al (2011) Robin sequence: from diagnosis to development of an effective management plan. Pediatrics 127:936–948. https://doi.org/10.1542/peds.2010-2615
Yasuda H, Do OC, Chen D et al (2017) A novel regulatory mechanism of type II collagen expression via a SOX9-dependent enhancer in intron 6. J Biol Chem 292:528–538. https://doi.org/10.1074/jbc.M116.758425
Lefebvre V, Huang W, Harley VR et al (1997) SOX9 is a potent activator of the chondrocyte-specific enhancer of the pro alpha1 (II) collagen gene. Mol Cell Biol 17:2336–2346. https://doi.org/10.1128/mcb.17.4.2336
Ni Q, Tan Y, Zhang X et al (2015) Prenatal ethanol exposure increases osteoarthritis susceptibility in female rat offspring by programming a low-functioning IGF-1 signaling pathway. Sci Rep 5:1–13. https://doi.org/10.1038/srep14711
Zhou G, Zheng Q, Engin F et al (2006) Dominance of SOX9 function over RUNX2 during skeletogenesis. Proc Natl Acad Sci U S A 103:19004–19009. https://doi.org/10.1073/pnas.0605170103
El Bouchikhi I, Belhassan K, Moufid FZ et al (2016) Noonan syndrome-causing genes: molecular update and an assessment of the mutation rate. Int J Pediatr Adolesc Med 3:133–142. https://doi.org/10.1016/j.ijpam.2016.06.003
Sinkala M, Nkhoma P, Mulder N, Martin DP (2021) Integrated molecular characterisation of the MAPK pathways in human cancers reveals pharmacologically vulnerable mutations and gene dependencies. Commun Biol 4. https://doi.org/10.1038/s42003-020-01552-6
Rauen KA (2013) The RASopathies. Annu Rev Genomics Hum Genet 14:355–369. https://doi.org/10.1146/annurev-genom-091212-153523
Bhambhani V, Muenke M (2014) Noonan syndrome. Am Fam Physician 89:37–43
Shaw AC, Kalidas K, Crosby AH et al (2007) The natural history of Noonan syndrome: a long-term follow-up study. Arch Dis Child 92:128–132. https://doi.org/10.1136/adc.2006.104547
Tidyman WE, Rauen KA (2009) The RASopathies: developmental syndromes of Ras/MAPK pathway dysregulation. Curr Opin Genet Dev 19:230–236. https://doi.org/10.1016/j.gde.2009.04.001
Tartaglia M, Mehler EL, Goldberg R et al (2001) Mutations in PTPN11, encoding the protein tyrosine phosphatase SHP-2, cause Noonan syndrome. Nat Genet 29:465–468. https://doi.org/10.1038/ng772
Sarkozy A, Digilio MC, Dallapiccola B (2008) Leopard syndrome. Orphanet J Rare Dis 3:1–8. https://doi.org/10.1186/1750-1172-3-13
Nava C, Hanna N, Michot C et al (2007) Cardio-facio-cutaneous and Noonan syndromes due to mutations in the RAS/MAPK signalling pathway: genotype-phenotype relationships and overlap with Costello syndrome. J Med Genet 44:763–771. https://doi.org/10.1136/jmg.2007.050450
Nishi E, Mizuno S, Nanjo Y et al (2015) A novel heterozygous MAP2K1 mutation in a patient with Noonan syndrome with multiple lentigines. Am J Med Genet Part A 167:407–411. https://doi.org/10.1002/ajmg.a.36842
Schmidt J, Ramis-Zaldivar JE, Nadeu F et al (2017) Mutations of MAP2K1 are frequent in pediatric-type follicular lymphoma and result in ERK pathway activation. Blood 130:323–327. https://doi.org/10.1182/blood-2017-03-776278
Allanson JE, Bohring A, Dorr H-G, Dufke A (2010) The face of Noonan syndrome: does phenotype predict genotype. Am J Med Genet A 152A:1960–1966. https://doi.org/10.1002/ajmg.a.33518
Jaouadi H, Ben CA, Kraoua L et al (2019) A severe clinical phenotype of Noonan syndrome with neonatal hypertrophic cardiomyopathy in the second case worldwide with RAF1 S259Y neomutation. Genet Res (Camb) 101:e6. https://doi.org/10.1017/S0016672319000041
Dinsmore CJ, Soriano P (2018) MAPK and PI3K signaling: at the crossroads of neural crest development. Dev Biol 444:S79–S97. https://doi.org/10.1016/j.ydbio.2018.02.003
Parada C, Han D, Grimaldi A et al (2015) Disruption of the ERK/MAPK pathway in neural crest cells as a potential cause of Pierre Robin sequence. Co Biol 142:3734–3745. https://doi.org/10.1242/dev.125328
Nakamura T, Gulick J, Pratt R, Robbins J (2009) Noonan syndrome is associated with enhanced pERK activity, the repression of which can prevent craniofacial malformations. PNAS 106:15436–15441
Newbern J, Zhong J, Wickramasinghe SR et al (2008) Mouse and human phenotypes indicate a critical conserved role for ERK2 signaling in neural crest development. PNAS 105:17115–17120
Lipinski RJ, Hammond P, O’Leary-Moore SK et al (2012) Ethanol-induced face-brain dysmorphology patterns are correlative and exposure-stage dependent. PLoS One. https://doi.org/10.1371/journal.pone.0043067
Higashiyama D, Saitsu H, Komada M et al (2007) Sequential developmental changes in holoprosencephalic mouse embryos exposed to ethanol during the gastrulation period. Birth Defects Res Part A Clin Mol Teratol 79:513–523. https://doi.org/10.1002/bdra.20367
Krantz ID, McCallum J, DeScipio C, Kaur M (2004) Cornelia de Lange syndrome is caused by mutations in NIPBL, the human homolog of Drosophila melanogaster Nipped-B. Nat Genet 36:631–635. https://doi.org/10.1038/ng1364.Cornelia
Tonkin ET, Wang TJ, Lisgo S et al (2004) NIPBL, encoding a homolog of fungal Scc2-type sister chromatid cohesion proteins and fly Nipped-B, is mutated in Cornelia de Lange syndrome. Nat Genet 36:636–641. https://doi.org/10.1038/ng1363
Kline AD, Moss JF, Selicorni A et al (2018) Diagnosis and management of Cornelia de Lange syndrome: first international consensus statement. Nat Rev Genet 19:649–666. https://doi.org/10.1038/s41576-018-0031-0
Kaur M, Descipio C, Mccallum J et al (2005) Precocious sister chromatid separation (PSCS) in Cornelia de Lange syndrome. Am J Med Genet A 138:27–31. https://doi.org/10.1002/ajmg.a.30919.Precocious
Rollins RA, Morcillo P, Dorsett D (1999) Nipped-B, a Drosophila homologue of chromosomal adherins, participates in activation by remote enhancers in the cut and Ultrabithorax genes. Genetics 152:577–593. https://doi.org/10.1093/genetics/152.2.577
Jahnke P, Xu W, Wülling M et al (2008) The Cohesin loading factor NIPBL recruits histone deacetylases to mediate local chromatin modifications. Nucleic Acids Res 36:6450–6458. https://doi.org/10.1093/nar/gkn688
Bhuiyan ZA, Klein M, Hammond P et al (2006) Genotype-phenotype correlations of 39 patients with Cornelia de Lange syndrome: the Dutch experience. J Med Genet 43:568–575. https://doi.org/10.1136/jmg.2005.038240
Selicorni A, Russo S, Gervasini C et al (2007) Clinical score of 62 Italian patients with Cornelia de Lange syndrome and correlations with the presence and type of NIPBL mutation. Clin Genet 72:98–108. https://doi.org/10.1111/j.1399-0004.2007.00832.x
Ansari M, Poke G, Ferry Q et al (2014) Genetic heterogeneity in Cornelia de Lange syndrome (CdLS) and CdLS-like phenotypes with observed and predicted levels of mosaicism. J Med Genet 51:659–668. https://doi.org/10.1136/jmedgenet-2014-102573
He J, Gu L, Zhang H, Zhou M (2011) Crosstalk between MYCN and MDM2-p53 signal pathways regulates tumor cell growth and apoptosis in neuroblastoma. Cell Cycle 10:2994–3002. https://doi.org/10.4161/cc.10.17.17118
Santos R, Kawauchi S, Jacobs RE et al (2016) Conditional creation and rescue of Nipbl-deficiency in mice reveals multiple determinants of risk for congenital heart defects. PLoS Biol 14:1–31. https://doi.org/10.1371/journal.pbio.2000197
Kotch LE, Sulik KK (1992) Patterns of ethanol-induced cell death in the developing nervous system of mice; neural fold states through the time of anterior neural tube closure. Int J Devl Neurosci 10:273–279
Chudley AE, Conry J, Cook JL et al (2005) Canadian guidelines for FASD. C Can Med Assoc J 172:S1–S21
Motahari Z, Moody SA, Maynard TM, Lamantia A (2019) In the line-up: deleted genes associated with DiGeorge/22q11.2 deletion syndrome: are they all suspects? J Neurodev Disord 11:1–28
Morrow BE, Emanuel DMMBS, Vermeesch JR, Scambler PJ (2018) Molecular genetics of 22q11.2 deletion syndrome. Am J Med Genet 176A:2070–2081. https://doi.org/10.1002/ajmg.a.40504
Bonthius DJ, Goodlett CR, West JR (1988) Blood alcohol concentration and severity of microencephaly in neonatal rats depend on the pattern of alcohol administration. Alcohol 5:209–214. https://doi.org/10.1016/0741-8329(88)90054-7
Pierce DR, West JR (1986) Blood alcohol concentration: a critical factor for producing fetal alcohol effects. Alcohol 3:269–272
Yutzey KE (2010) DiGeorge syndrome, Tbx1, and retinoic acid signaling come full circle. Circ Res 106:630–632. https://doi.org/10.1161/CIRCRESAHA.109.215319
Maynard TM, Gopalakrishna D, Meechan DW et al (2013) 22q11 gene dosage establishes an adaptive range for sonic hedgehog and retinoic acid signaling during early development. Hum Mol Genet 22:300–312. https://doi.org/10.1093/hmg/dds429
Garg V, Yamagishi C, Hu T et al (2001) Tbx1, a DiGeorge syndrome candidate gene, is regulated by sonic hedgehog during pharyngeal arch development. Dev Biol 73:62–73. https://doi.org/10.1006/dbio.2001.0283
Karpinski BA, Maynard TM, Fralish MS et al (2014) Dysphagia and disrupted cranial nerve development in a mouse model of DiGeorge (22q11) deletion syndrome. Dis Model Mech 7:245–257. https://doi.org/10.1242/dmm.012484
Corsten-Janssen N, Saitta SC, Hoefsloot LH et al (2013) More clinical overlap between 22q11.2 deletion syndrome and charge syndrome than often anticipated. Mol Syndromol 4:235–245. https://doi.org/10.1159/000351127
Tazumi S, Yabe S, Uchiyama H (2010) Paraxial T-box genes, Tbx6 and Tbx1, are required for cranial chondrogenesis and myogenesis. Dev Biol 346:170–180. https://doi.org/10.1016/j.ydbio.2010.07.028
George RM, Firulli AB (2021) Epigenetics and heart development. Front Cell Dev Biol 9:1–10. https://doi.org/10.3389/fcell.2021.637996
Keverne EB (2015) Genomic imprinting, action, and interaction of maternal and fetal genomes. Proc Natl Acad Sci U S A 112:6834–6840. https://doi.org/10.1073/pnas.1411253111
Forstner AJ, Degenhardt F, Schratt G, Nöthen MM (2013) MicroRNAs as the cause of schizophrenia in 22q11.2 deletion carriers, and possible implications for idiopathic disease: a mini-review. Front Mol Neurosci 6:1–10. https://doi.org/10.3389/fnmol.2013.00047
Suzuki HI, Young RA, Sharp PA (2017) Super-enhancer-mediated RNA processing revealed by integrative microRNA network analysis. Cell 168:1000–1014.e15. https://doi.org/10.1016/j.cell.2017.02.015
Napoli C, Schiano C, Soricelli A (2019) Increasing evidence of pathogenic role of the Mediator (MED) complex in the development of cardiovascular diseases. Biochimie 165:1–8. https://doi.org/10.1016/j.biochi.2019.06.014
Huang P, Nedelcu D, Watanabe M et al (2016) Cellular cholesterol directly activates Smoothened in Hedgehog signaling. Cell 166:1176–1187. https://doi.org/10.1016/j.cell.2016.08.003.Cellular
Che Y, Siprashvili Z, Kovalski JR et al (2019) KRAS regulation by small non-coding RNAs and SNARE proteins. Nat Commun 10. https://doi.org/10.1038/s41467-019-13106-4
Motta M, Fidan M, Bellacchio E et al (2019) Dominant Noonan syndrome-causing LZTR1 mutations specifically affect the Kelch domain substrate-recognition surface and enhance RAS-MAPK signaling. Hum Mol Genet 28:1007–1022. https://doi.org/10.1093/hmg/ddy412
Abe T, Umeki I, Kanno SI et al (2020) LZTR1 facilitates polyubiquitination and degradation of RAS-GTPases. Cell Death Differ 27:1023–1035. https://doi.org/10.1038/s41418-019-0395-5
Bigenzahn JW, Collu GM, Kartnig F et al (2018) LZTR1 is a regulator of RAS ubiquitination and signaling. Science (80-) 362:1171–1177. https://doi.org/10.1126/science.aap8210
Castel P, Cheng A, Cuevas-Navarro A et al (2019) RIT1 oncoproteins escape LZTR1-mediated proteolysis. Science (80-) 363:1226–1230. https://doi.org/10.1126/science.aav1444
Cuevas-Navarro A, Van R, Cheng A et al (2021) The RAS GTPase RIT1 compromises mitotic fidelity through spindle assembly checkpoint suppression. Curr Biol 31:3915–3924.e9. https://doi.org/10.1016/j.cub.2021.06.030
Moutin E, Nikonenko I, Stefanelli T et al (2017) Palmitoylation of cdc42 promotes spine stabilization and rescues spine density deficit in a mouse model of 22q11.2 deletion syndrome. Cereb Cortex 27:3618–3629. https://doi.org/10.1093/cercor/bhw183
Cooper MK, Wassif CA, Krakowiak PA et al (2003) A defective response to Hedgehog signaling in disorders of cholesterol biosynthesis. Nat Genet 33:508–513. https://doi.org/10.1038/ng1134
Alvarez JI, Dodelet-Devillers A, Kebir H et al (2011) The hedgehog pathway promotes blood-brain barrier integrity and CNS immune quiescence. Science (80-) 334:1727–1731. https://doi.org/10.1126/science.1206936
Liebner S, Corada M, Bangsow T et al (2008) Wnt/β-catenin signaling controls development of the blood – brain barrier. J Cell Biol 183:409–417. https://doi.org/10.1083/jcb.200806024
Napoli E, Tassone F, Wong S et al (2015) Mitochondrial citrate transporter-dependent metabolic signature in the 22q11.2 deletion syndrome. J Biol Chem 290:23240–23253. https://doi.org/10.1074/jbc.M115.672360
Kohn AD, Moon RT (2005) Wnt and calcium signaling: β-catenin-independent pathways. Cell Calcium 38:439–446. https://doi.org/10.1016/j.ceca.2005.06.022
Khan TA, Revah O, Gordon A et al (2020) Neuronal defects in a human cellular model of 22q11.2 deletion syndrome. Nat Med 26(12):1888–1898. Springer US
Gao L, Gorski JL, Chen CS (2011) The Cdc42 guanine nucleotide exchange factor FGD1 regulates osteogenesis in human mesenchymal stem cells. Am J Pathol 178:969–974. https://doi.org/10.1016/j.ajpath.2010.11.051
Rangamani P, Levy MG, Khan S, Oster G (2016) Paradoxical signaling regulates structural plasticity in dendritic spines. Proc Natl Acad Sci U S A 113:E5298–E5307. https://doi.org/10.1073/pnas.1610391113
Umeki I, Niihori T, Abe T et al (2019) Delineation of LZTR1 mutation-positive patients with Noonan syndrome and identification of LZTR1 binding to RAF1–PPP1CB complexes. Hum Genet 138:21–35. https://doi.org/10.1007/s00439-018-1951-7
Moon AM, Guris DL, Seo J et al (2006) Crkl deficiency disrupts Fgf8 signaling in a mouse model of 22q11 deletion syndromes. Dev Cell 10:71–80. https://doi.org/10.1016/j.devcel.2005.12.003
Zhu C, Chen C, Huang J et al (2015) SUMOylation at K 707 of DGCR8 controls direct function of primary microRNA. Nucleic Acids Res 43:7945–7960. https://doi.org/10.1093/nar/gkv741
Herbert KM, Pimienta G, Degregorio SJ et al (2013) Article phosphorylation of DGCR8 increases its intracellular stability and induces a progrowth miRNA profile. Cell Rep 5:1070–1081. https://doi.org/10.1016/j.celrep.2013.10.017
Samuels IS, Karlo JC, Faruzzi AN et al (2008) Deletion of ERK2 mitogen-activated protein kinase identifies its key roles in cortical neurogenesis and cognitive function. J Neurosci 28:6983–6995. https://doi.org/10.1523/JNEUROSCI.0679-08.2008
Badodi S, Dubuc A, Zhang X et al (2017) Convergence of BMI1 and CHD7 on ERK signaling in convergence of BMI1 and CHD7 on ERK signaling in medulloblastoma. Cell Rep 21:2772–2784. https://doi.org/10.1016/j.celrep.2017.11.021
Jamadagni P, Breuer M, Schmeisser K et al (2021) Chromatin remodeller CHD7 is required for GABAergic neuron development by promoting PAQR 3 expression. EMBO Rep 22:1–18. https://doi.org/10.15252/embr.202050958
Martinelli S, Krumbach OHF, Pantaleoni F et al (2018) Functional dysregulation of CDC42 causes diverse developmental phenotypes. Am J Hum Genet 102:309–320. https://doi.org/10.1016/j.ajhg.2017.12.015
Deroanne CF, Hamelryckx D, Ho TTG et al (2005) Cdc42 downregulates MMP-1 expression by inhibiting the ERK1/2 pathway. J Cell Sci 118:1173–1183. https://doi.org/10.1242/jcs.01707
Filigheddu N, Gnocchi VF, Coscia M et al (2007) Gene targeting of Cdc42 and Cdc42GAP affirms the critical involvement of Cdc42 in filopodia induction, directed migration, and proliferation in primary mouse embryonic fibroblasts. Mol Biol Cell 18:986–994. https://doi.org/10.1091/mbc.E06
Villalobo A, Berchtold MW (2020) The role of calmodulin in tumor cell migration, invasiveness, and metastasis. Int J Mol Sci 21. https://doi.org/10.3390/ijms21030765
Daniel EE, Cho WJ (2006) Caveolae and calcium handling, a review and a hypothesis. J Cell Mol Med 10:529–544
Furuchi T, Anderson RGW (1998) Cholesterol depletion of Caveolae causes hyperactivation of extracellular signal-related kinase (ERK)*. J Biol Chem 273:21099–21104. https://doi.org/10.1074/jbc.273.33.21099
Park JH, Han HJ (2009) Caveolin-1 plays important role in EGF-induced migration and proliferation of mouse embryonic stem cells: involvement of PI3K/Akt and ERK. Am J Physiol Cell Physiol 297:935–944. https://doi.org/10.1152/ajpcell.00121.2009
Shikanai M, Yoshiaki V, Shikanai M et al (2018) Caveolin-1 promotes early neuronal maturation via caveolae-independent trafficking of N- cadherin and L1 Caveolin-1 promotes early neuronal maturation via caveolae-independent trafficking of N-cadherin and L1. iScience 7:53–67. https://doi.org/10.1016/j.isci.2018.08.014
Parton RG, Simons K (2007) The multiple faces of caveolae. Nat Rev 8:185–194. https://doi.org/10.1038/nrm2122
Wang P, Weng J, Anderson RGW (2005) OSBP is a cholesterol-regulated scaffolding protein in control of ERK1/2 activation. Science (80-) 307:1472–1477
Grande-garcía A, Echarri A, De Rooij J et al (2007) Caveolin-1 regulates cell polarization and directional migration through Src kinase and rho GTPases. J Cell Biol 177:683–694. https://doi.org/10.1083/jcb.200701006
Mao H, Diehl AM, Li YX (2009) Sonic hedgehog ligand partners with caveolin-1 for intracellular transport. Lab Investig 89:290–300. https://doi.org/10.1038/labinvest.2008.163
Dou X, Wilkemeyer MF, Menkari CE et al (2013) Mitogen-activated protein kinase modulates ethanol inhibition of cell adhesion mediated by the L1 neural cell adhesion molecule. PNAS 110:5683–5688. https://doi.org/10.1073/pnas.1221386110
Lussier AA, Morin AM, MacIsaac JL et al (2018) DNA methylation as a predictor of fetal alcohol spectrum disorder. Clin Epigenetics 10:5. https://doi.org/10.1186/s13148-018-0439-6
Lussier AA, Bodnar TS, Weinberg J (2021) Intersection of epigenetic and immune alterations: implications for fetal alcohol spectrum disorder and mental health. Front Neurosci 15. https://doi.org/10.3389/fnins.2021.788630
Wallén E, Auvinen P, Kaminen-Ahola N (2021) The effects of early prenatal alcohol exposure on epigenome and embryonic development. Genes (Basel) 12. https://doi.org/10.3390/genes12071095
Alberry B, Laufer BI, Chater-Diehl E, Singh SM (2021) Epigenetic impacts of early life stress in fetal alcohol spectrum disorders shape the neurodevelopmental continuum. Front Mol Neurosci 14:1–17. https://doi.org/10.3389/fnmol.2021.671891
Jongmans MCJ, Hoefsloot LH, Van Der Donk KP et al (2008) Familial CHARGE syndrome and the CHD7 gene: a recurrent missense mutation, intrafamilial recurrence and variability. Am J Med Genet 146A:43–50. https://doi.org/10.1002/ajmg.a
Pauli S, Bajpai R, Borchers A (2017) CHARGEd with neural crest defects. Am J Med Genet 175C:478–486. https://doi.org/10.1002/ajmg.c.31584
Feng W, Shao C, Liu H (2017) Versatile roles of the chromatin remodeler CHD7 during brain development and disease. Front Mol Neurosci 10:1–8. https://doi.org/10.3389/fnmol.2017.00309
Blake KD, Prasad C (2006) CHARGE syndrome. Orphanet J Rare Dis 1:1–8. https://doi.org/10.1186/1750-1172-1-34
Liu Y, Balaraman Y, Wang G et al (2009) Alcohol exposure alters DNA methylation profiles in mouse embryos at early neurulation. Epigenetics 4:500–511. https://doi.org/10.4161/epi.4.7.9925
Resendiz M, Chen Y, Öztürk NC, Zhou FC (2013) Epigenetic medicine and fetal alcohol spectrum disorders. Epigenomics 5:73–86. https://doi.org/10.2217/epi.12.80
Guo Y, Monahan K, Wu H et al (2012) CTCF/cohesin-mediated DNA looping is required for protocadherin α promoter choice. Proc Natl Acad Sci U S A 109:21081–21086. https://doi.org/10.1073/pnas.1219280110
Remeseiro S, Cuadrado A, Gómez-Lãpez G et al (2012) A unique role of cohesin-SA1 in gene regulation and development. EMBO J 31:2090–2102. https://doi.org/10.1038/emboj.2012.60
Laufer BI, Kapalanga J, Castellani CA et al (2015) Associative DNA methylation changes in children with prenatal alcohol exposure. Epigenomics 7:1259–1274
Laufer BI, Chater-Diehl EJ, Kapalanga J, Singh SM (2017) Long-term alterations to DNA methylation as a biomarker of prenatal alcohol exposure: from mouse models to human children with fetal alcohol spectrum disorders. Alcohol 60:67–75. https://doi.org/10.1016/j.alcohol.2016.11.009
Cook JL, Green CR, Lilley CM et al (2016) Fetal alcohol spectrum disorder: a guideline for diagnosis across the lifespan. CMAJ 188:191–197
Popova S, Lange S, Shield K et al (2016) Comorbidity of fetal alcohol spectrum disorder: a systematic review and meta-analysis. Lancet 387:978–987. https://doi.org/10.1016/S0140-6736(15)01345-8
Denny LA, Coles S, Blitz R (2017) Fetal alcohol syndrome and fetal alcohol Spectrum disorders. Am Fam Physician 96:515–522. https://doi.org/10.1002/0471695998.mgs020
Leibson T, Neuman G, Chudley AE, Koren G (2014) The differential diagnosis of fetal alcohol spectrum disorder. J Popul Ther Clin Pharmacol 21:1–30
Blanck-Lubarsch M, Dirksen D, Feldmann R et al (2019) Tooth malformations, dmft index, speech impairment and oral habits in patients with fetal alcohol syndrome. Int J Environ Res Public Health 16:1–12. https://doi.org/10.3390/ijerph16224401
Blanck-Lubarsch M, Dirksen D, Feldmann R et al (2020) Children with fetal alcohol syndrome (FAS): 3D-analysis of palatal depth and 3D-metric facial length. Int J Environ Res Public Health 17. https://doi.org/10.3390/ijerph17010095
Mcdonald-McGinn DM, Hain HS, Emanuel BS, Zackai EH (2021) 22q11.2 deletion syndrome. Nat Rev Dis Primers 1:15071
Lewyllie A, Roosenboom J, Indencleef K et al (2017) A comprehensive craniofacial study of 22q11.2 deletion syndrome. J Dent Res 96:1386–1392. https://doi.org/10.1177/0022034517720630
Roberts AE, Allanson JE, Tartaglia M, Gelb BD (2013) Noonan syndrome. Lancet 381:333–342. https://doi.org/10.1016/S0140-6736(12)61023-X
Unger S, Scherer G, Superti-Furga A (2008) Campomelic dysplasia. In: GeneReviews. https://www.ncbi.nlm.nih.gov/books/NBK1760/
Corbani S, Chouery E, Eid B et al (2011) Mild campomelic dysplasia: report on a case and review. Mol Syndromol 1:163–168. https://doi.org/10.1159/000322861
Roberts GL, Beiraghi S (1989) Dental abnormalities associated with campomelic syndrome: case report. Pediatr Dent 11:43–46
Kline AD, Krantz ID, Sommer A et al (2007) Cornelia de Lange syndrome: clinical review, diagnostic and scoring systems, and anticipatory guidance. Am J Med Genet Part A Genet 143A:1287–1296. https://doi.org/10.1002/ajmg.a
Kelley RI, Hennekam RCM (2000) The Smith-Lemli-Opitz syndrome. 321–335
Anthony B, Vinci-Booher S, Wetherill L et al (2010) Alcohol-induced facial dysmorphology in C57BL/6 mouse models of fetal alcohol spectrum disorder. Alcohol 44:659–671. https://doi.org/10.1016/j.alcohol.2010.04.002
Kaminen-ahola N, Fahey P, Cox TC et al (2010) Maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model maternal ethanol consumption alters the epigenotype and the phenotype of offspring in a mouse model. PLoS One 6:1–10. https://doi.org/10.1371/journal.pgen.1000811
Sowell ER, Mattson SN, Kan E et al (2008) Abnormal cortical thickness and brain-- behavior correlation patterns in individuals with heavy prenatal alcohol exposure. Cereb Cortex 18:136–144. https://doi.org/10.1093/cercor/bhm039
Jarmasz JS, Basalah DA, Chudley AE, Del Bigio MR (2017) Human brain abnormalities associated with prenatal alcohol exposure and fetal alcohol spectrum disorder. Neurophathol Exp Neurol 76:813–833. https://doi.org/10.1093/jnen/nlx064
Coulter C, Leech R, Schaefer B et al (1993) Midline cerebral dysgenesis, dysfunction of the hypothalamic-pituitary axis, and fetal alcohol effects. Arch Neurol 50:771–775
Strömland K, Pinazo-Durán DM (2002) Invited review ophthalmic involvement in the fetal alcohol syndrome: clinical and animal model studies. Alcohol Alcohol 37:2–8
Church MW, Eldis F, Blakley BW, Bawle EV (1997) Hearing, language, speech, vestibular, and dentofacial disorders in fetal alcohol syndrome. Alcohol Clin Exp Res 21:227–237
Peiffer J, Majweski F, Fischbach H et al (1979) Alcohol embryo- and fetopathy: neuropathology of 3 children and 3 fetuses. J Neurol Sci 41:125–137
Kar P, Reynolds JE, Grohs MN et al (2021) White matter alterations in young children with prenatal alcohol exposure. Dev Neurobiol 00:1–11. https://doi.org/10.1002/dneu.22821
Ghazi F, Mohammad S, Aarabi H, Haghshomar M (2019) White matter microstructure in fetal alcohol spectrum disorders: a systematic review of diffusion tensor imaging studies. Hum Brain Mapp 40:1017–1036. https://doi.org/10.1002/hbm.24409
Hultman SC, Riski JE, Cohen SR et al (2000) Chiari malformation, cervical spine anomalies, and neurologic deficits in velocardiofacial syndrome. Plast Reconstr Surg 106:16–24
Campbell LE, Daly E, Toal F et al (2006) Brain and behaviour in children with 22q11.2 deletion syndrome: a volumetric and voxel-based morphometry MRI study. Brain 129:1218–1228. https://doi.org/10.1093/brain/awl066
Villalón-Reina J, Martínez K, Qu X et al (2020) Altered white matter microstructure in 22q11.2 deletion syndrome: a multisite diffusion tensor imaging study. Mol Psychiatry 25:2818–2831. https://doi.org/10.1038/s41380-019-0450-0
Rogdaki M, Gudbrandsen M, Mccutcheon RA, et al (2020) Magnitude and heterogeneity of brain structural abnormalities in 22q11.2 deletion syndrome: a meta-analysis. Mol Psychiatry 1704–1717. https://doi.org/10.1038/s41380-019-0638-3
Fattah M, Raman MM, Reiss AL, Green T (2021) PTPN11 mutations in the Ras-MAPK signaling pathway affect human white matter microstructure. Cereb Cortex 31:1489–1499. https://doi.org/10.1093/cercor/bhaa299
Amico AD, Cipullo MB, Falco M et al (2021) Clinical report of a brain magnetic resonance imaging finding in Noonan syndrome. Childs Nerv Syst 37:3963–3966
Carcavilla A, Suárez-ortega L, Rodríguez A et al (2020) Noonan syndrome: genetic and clinical update and treatment options. An Pediatr 93. https://doi.org/10.1016/j.anpede.2020.04.009
Allanson JE, Roberts AE (2019) Noonan syndrome. In: GeneReviews, pp 1–36
Hartill VL, Dillon MW, Warren DJ, Blyth M (2017) RAF1 -associated Noonan syndrome presenting antenatally with an abnormality of skull shape, subdural haematoma and associated with novel cerebral malformations. Clin Dysmorphol 26:101–106. https://doi.org/10.1097/MCD.0000000000000153
Shiohama T, Mcdavid J, Levman J, Takahashi E (2019) NeuroImage: clinical quantitative brain morphological analysis in CHARGE syndrome. NeuroImage Clin 23:101866. https://doi.org/10.1016/j.nicl.2019.101866
van Ravenswaaij-Arts CM, Hefner M, Blake K, Martin DM (2020) CHD7 disorder summary genetic counseling GeneReview scope diagnosis suggestive findings. In: GeneReviews, pp 1–31
Lyonnet S, Rappaport R, Netchine I (2005) CHARGE syndrome includes hypogonadism and abnormal olfactory bulb development. J Clin Endocrinol Metab 90:5621–5626. https://doi.org/10.1210/jc.2004-2474
Hoch MJ, Patel SH, Jethanamest D et al (2017) Head and neck MRI findings in CHARGE syndrome. AJNR Am J Neuroradiol 38:2357–2363
Mansour S, Offiah AC, McDowall S et al (2002) The phenotype of survivors of campomelic dysplasia. J Med Genet 39:597–602. https://doi.org/10.1136/jmg.39.8.597
Matsumoto A, Imagawa E, Miyake N et al (2018) The presence of diminished white matter and corpus callosal thinning in a case with a SOX9 mutation. Brain and Development 40:325–329. https://doi.org/10.1016/j.braindev.2017.09.002
Whitehead MT, Nagaraj UD, Pearl PL (2015) Neuroimaging features of Cornelia de Lange syndrome. 1198–1205. https://doi.org/10.1007/s00247-015-3300-5
Bottani A, Orrico A, Galli L, Karam O (2007) Unilateral focal Polymicrogyria in a patient with classical Aarskog – Scott syndrome due to a novel missense mutation in an evolutionary conserved RhoGEF domain of the faciogenital dysplasia gene FGD1. Am J Med Genet Part A 143A:2334–2338. https://doi.org/10.1002/ajmg.a
Shalev SA, Chervinski E, Weiner E et al (2006) Clinical variation of Aarskog syndrome in a large family with 2189delA in the FGD1 gene. Am J Med Genet 140A:162–165. https://doi.org/10.1002/ajmg.a
Malgorzata S, Nowaczyk JM (2021) Smith-Lemli-Opitz syndrome
Lee RWY, Conley SK, Gropman A et al (2013) Brain magnetic resonance imaging findings in smith-lemli-opitz syndrome. Am J Med Genet Part A 161:2407–2419. https://doi.org/10.1002/ajmg.a.36096
Maroni G (2022) X-linked opitz G/BBB syndrome
Graham JM Jr, Visootsak J, Dykens E et al (2008) Behavior of 10 patients with FG syndrome (Opitz – Kaveggia syndrome) and the p.R961W mutation in the MED12 gene. Am J Med Genet Part A 146A:3011–3017. https://doi.org/10.1002/ajmg.a.32553
Ozonoff S, Williams BJ, Rauch AM, Opitz JM (2000) Behavior phenotype of FG syndrome: cognition, personality, and behavior in eleven affected boys. Am J Med Genet 97:112–118
Cao W, Li W, Han H et al (2014) Prenatal alcohol exposure reduces magnetic susceptibility contrast and anisotropy in the white matter of mouse brains. NeuroImage. https://doi.org/10.1016/j.neuroimage.2014.08.035
Stevens SA, Nash K, Koren G, Rovet J (2013) Autism characteristics in children with fetal alcohol spectrum disorders. Child Neuropsychol 19:579–587. https://doi.org/10.1080/09297049.2012.727791
Goril S, Zalai D, Scott L, Shapiro CM (2016) Sleep and melatonin secretion abnormalities in children and adolescents with fetal alcohol spectrum disorders. Sleep Med 23:59–64. https://doi.org/10.1016/j.sleep.2016.06.002
Hellemans KG, Verma P, Yoon E et al (2010) Prenatal alcohol exposure and chronic mild stress differentially alter depressive- and anxiety-like behaviors in male and female offspring. Alcohol Clin Exp Res 34:633–645
Pierpont EI (2016) Neuropsychological functioning in individuals with Noonan syndrome: a systematic literature review with educational and treatment recommendations. J Pediatr Neuropsychol 14–33. https://doi.org/10.1007/s40817-015-0005-5
Hartshorne TS, Stratton KK, Brown D, Schmittel SMMC (2017) Behavior in CHARGE syndrome. Am J Med Genet 175C:431–438. https://doi.org/10.1002/ajmg.c.31588
Hartshorne TS, Cypher AD (2004) Challenging behavior in CHARGE syndrome timothy. Ment Heal Asp Dev Disabil 7:41–52
Wulffaert J, Scholte EM, Dijkxhoorn YM, Van Berckelaer-onnes IA (2009) Parenting stress in CHARGE syndrome and the relationship with child characteristics. J Dev Phys Disabil 21:301–313. https://doi.org/10.1007/s10882-009-9143-y
Lee JJ, Thottam PJ, Ford MD, Jabbour N (2015) Characteristics of sleep apnea in infants with Pierre-Robin sequence: is there improvement with advancing age? Int J Pediatr Otorhinolaryngol 79:2059–2067. https://doi.org/10.1016/j.ijporl.2015.09.014
Giani L, Michelini G, Nobile M et al (2022) Behavioral markers of social anxiety in Cornelia de Lange syndrome: a brief systematic review. J Affect Disord 299:636–643. https://doi.org/10.1016/j.jad.2021.12.099
Kaname T, Yanagi K, Okamoto N, Naritomi K (2006) Neurobehavioral disorders in patients with Aarskog – Scott syndrome affected by novel FGD1 mutations. Am J Med Genet 140A:1331–1332. https://doi.org/10.1002/ajmg.a
Hamzeh AR, Saif F, Nair P, et al (2017) A novel, putatively null, FGD1 variant leading to Aarskog-Scott syndrome in a family from UAE. BMC Pediatr 4–9. https://doi.org/10.1186/s12887-017-0781-4
El Shawa H, Abbott CW, Huffman KJ (2013) Prenatal ethanol exposure disrupts intraneocortical circuitry, cortical gene expression, and behavior in a mouse model of FASD. J Neurosci 33:18893–18905. https://doi.org/10.1523/JNEUROSCI.3721-13.2013
Kleiber ML, Mantha K, Stringer RL, Singh SM (2013) Neurodevelopmental alcohol exposure elicits long-term changes to gene expression that alter distinct molecular pathways dependent on timing of exposure. J Neurodev Disord 5:6. https://doi.org/10.1186/1866-1955-5-6
Kleiber ML, Wright E, Singh SM (2011) Maternal voluntary drinking in C57BL/6J mice: advancing a model for fetal alcohol spectrum disorders. Behav Brain Res 223:376–387. https://doi.org/10.1016/j.bbr.2011.05.005
Caldwell KK, Sheema S, Paz RD et al (2008) Fetal alcohol spectrum disorder-associated depression: evidence for reductions in the levels of brain-derived neurotrophic factor in a mouse model. Pharmacol Biochem Behav 90:614–624. S0091-3057(08)00155-X [pii]. https://doi.org/10.1016/j.pbb.2008.05.004
Kim M, Kogan N, Slack FJ (2016) Cis-acting elements in its 3’ UTR mediate post-transcriptional regulation of KRAS. Oncotarget 7:11770–11784. https://doi.org/10.18632/oncotarget.7599
Acknowledgments
We wish to thank Abraham Fainsod, Songyan Liu, and Molly Pind for enlightening and provocative discussions over the years that contributed to this manuscript.
Funding
This work was funded in part by grants to GGH from the Canadian Institutes of Health Research (PJT-165847), Manitoba Liquor & Lotteries Corporation (55201 and 55380), and Kids Brain Health Network (48694) and to LM from the Kids Brain Health Network (54525).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
McKay, L., Petrelli, B., Chudley, A.E., Hicks, G.G. (2022). Genetics of FASD: Confounding Rare Craniofacial and Neurodevelopmental Disorders May Identify Ethanol-Sensitizing Genetic Variants of FASD. In: Chudley, A.E., Hicks, G.G. (eds) Fetal Alcohol Spectrum Disorder. Neuromethods, vol 188. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2613-9_5
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2613-9_5
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2612-2
Online ISBN: 978-1-0716-2613-9
eBook Packages: Springer Protocols