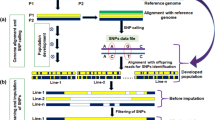
Abstract
Marker-assisted selection (MAS) plays a pivotal role in a breeding program where molecular DNA markers are used for phenotypic selections in crop improvement. Several markers have been used where SNPs (single-nucleotide polymorphisms) have been identified and effectively used. Next-generation sequencing (NGS) technologies have made significant changes to whole-genome sequencing revolutionizing plant breeding. Genotype by sequencing (GBS) is a rapid, cost-effective, and high-throughput method in NGS which enables genotyping of large populations with the discovery of SNPs. The GBS approach includes digestion of genomic DNA with restriction enzymes followed by ligation of barcode adapters, PCR amplification, and sequencing of the amplified DNA pool on a single lane of flow cells. This method has been developed and applied in the sequencing of multiplexed genomic samples. GBS is implemented successfully in genome-wide association study (GWAS), diversity studies, QTL mapping, genetic linkage analysis, marker discovery, and genomic selection under large-scale plant breeding programs.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of a genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Melake-Berhan A, Hulbert SH, Butler LG, Bennetzen JL (1993) Structure and evolution of the genomes of Sorghum bicolor and Zea mays. Theor Appl Genet 86:598–604
Ejeta G, Knoll JE (2007) Marker-assisted selection in sorghum. In: Genomics-assisted crop improvement, vol 2. Springer, New York, pp 187–205. https://doi.org/10.1007/978-1-4020-6297-1
Williams JG, Kubelik AR, Livak KJ, Rafalski JA, Tingey SV (1990) DNA polymorphisms amplified by arbitrary primers are useful as genetic markers. Nucleic Acids Res 18(22):6531–6535
Paran I, Michelmore RW (1993) Development of reliable PCR-based markers linked to downy mildew resistance genes in lettuce. Theor Appl Genet 85:985–993. https://doi.org/10.1007/BF00215038
Konieczny A, Ausubel FM (1993) A procedure for mapping Arabidopsis mutations using co-dominant ecotype-specific PCR-based markers. Plant J 4:403–410. https://doi.org/10.1046/j.1365-313X.1993.04020403
Litt M, Luty JA (1986) A hypervariable microsatellite revealed by invitro amplification of a dinucleotide repeat within the cardiac muscle actin gene. Am J Hum Genet 44:397–401
Salimath SS, de Oliveira AC, Bennetzen J, Godwin ID (1995) Assessment of genomic origin and genetic diversity in the genus Eleusine with DNA markers. Genome 38:757–763. https://doi.org/10.1139/g95-096
Vos P, Hogers R, Bleeker M, Reijans M, Lee TVD, Hornes M, Friters A, Pot J, Paleman J, Kuiper M, Zabeau M (1995) AFLP: a new technique for DNA fingerprinting. Nucleic Acids Res 23(21):4407–4414
Desmarais D, Zhong Y, Chakraborty R, Perreault C, Busque L (1998) Development of a highly polymorphic STR marker for identity testing purposes at the human androgen receptor gene (HUMARA). J Forensic Sci 43(5):1046–1049
Wang DG, Fan JB, Siao CJ, Berno A, Young P et al (1998) Large-scale identification, mapping, and genotyping of single-nucleotide polymorphisms in the human genome. Science 280(5366):1077–1082
Getachew SE, Bille NH, Bell JM, Gebreselassie W (2019) Genotyping by sequencing for plant breeding- a review. Adv Biotechnol Microbiol 14:555891. https://doi.org/10.19080/AIBM.2019.14.555891
He J, Zhao X, Laroche A, Lu Z-X, Liu H, Li Z (2014) Genotyping-by-sequencing (GBS), an ultimate marker-assisted selection (MAS) tool to accelerate plant breeding. Front Plant Sci 5:1–8. https://doi.org/10.3389/fpls.2014.00484
Madhusudhana R (2019) Marker-assisted breeding in sorghum. In: Breeding sorghum for diverse end uses, pp 93–114. Woodhead Publishing. https://doi.org/10.1016/B978-0-08-101879-8.00006-1
Francia E, Tacconi G, Crosatti C, Barabaschi D, Bulgarelli D, Dall’Aglio E et al (2005) Marker assisted selection in crop plants. Plant Cell Tissue Organ Cult 82:317–342. https://doi.org/10.1007/s11240-005-2387-z
Yoshimura S, Yoshimura A, Iwata N, Mccouch SR, Abenes ML, Baraoidan MR, Mew TW, Nelson RJ (1995) Tagging and combining bacterial blight resistance genes in rice using RAPD and RFLP markers. Mol Breed 1:375–387
Thudi M, Li Y, Jackson SA, May GD, Varshney RK (2012) Current state-of-art of sequencing technologies for plant genomics research. Brief Funct Genomics 11:3–11. https://doi.org/10.1093/bfgp/elr045
Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG..., Roe PM (2008) Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456(7218):53–59. https://doi.org/10.1038/nature07517
Rothberg JM, Hinz W, Rearick TM, Schultz J, Mileski W, Davey M et al (2011) An integrated semiconductor device enabling non-optical genome sequencing. Nature 475:348–352. https://doi.org/10.1038/nature10242
Metzker ML (2010) Sequencing technologies–the next generation. Nat Rev Genet 11:31–46. https://doi.org/10.1038/nrg2626
Mardis ER (2008) Next-generation DNA sequencing methods. Annu Rev Genomics Hum Genet 9:387–402. https://doi.org/10.1146/annurev.genom.9.081307.164359
Elshire RJ, Glaubitz JC, Sun Q, Poland JA, Kawamoto K, Buckler ES et al (2011) A robust, simple genotyping-by-sequencing (GBS) approach for high diversity species. PLoS One 6:e19379. https://doi.org/10.1371/journal.pone.0019379
Beissinger TM, Hirsch CN, Sekhon RS, Foerster JM, Johnson JM, Muttoni G et al (2013) Marker density and read depth for genotyping populations using genotyping-by-sequencing. Genetics 193:1073–1081. https://doi.org/10.1534/genetics.112.147710
Heffelfinger C, Fragoso CA, Moreno MA, Overton JD, Mottinger JP, Zhao H et al (2014) Flexible and scalable genotyping-by-sequencing strategies for population studies. BMC Genomics 15:1–23. https://doi.org/10.1186/1471-2164-15-979
Davey JW, Hohenlohe PA, Etter PD, Boone JQ, Catchen JM, Blaxter ML (2011) Genome-wide genetic marker discovery and genotyping using next-generation sequencing. Nat Rev Genet 12:499–510. https://doi.org/10.1038/nrg3012
Ward JA, Bhangoo J, Fernández-Fernández F, Moore P, Swanson JD, Viola R et al (2013) Saturated linkage map construction in Rubus idaeus using genotyping by sequencing and genome-independent imputation. BMC Genomics 14:2. https://doi.org/10.1186/1471-2164-14-2
Poland JA, Brown PJ, Sorrells ME, Jannink JL (2012) Development of high-density genetic maps for barley and wheat using a novel two-enzyme genotyping-by-sequencing approach. PLoS One 7:e32253. https://doi.org/10.1371/journal.pone.0032253
Abebe KS (2019) Genotype by sequencing method and its application for crop improvement (a review). Adv Biosci Bioeng 7(1):1
Scheben A, Batley J, Edwards D (2017) Genotyping-by-sequencing approaches to characterize crop genomes: choosing the right tool for the right application. Plant Biotechnol J 15(2):149–161
Deschamps S, Llaca V, May GD (2012) Genotyping-by-sequencing in plants. Biology 1:460–483. https://doi.org/10.3390/biology1030460
Schneeberger K, Ossowski S, Lanz C, Juul T, Petersen AH, Nielsen KL et al (2009) SHORE map: simultaneous mapping and mutation identification by deep sequencing. Nat Methods 6:550–551. https://doi.org/10.1038/nmeth0809-550
Romay MC, Millard MJ, Glaubitz JC, Peiffer JA, Swarts KL, Casstevens TM et al (2013) Comprehensive genotyping of the USA national maize inbred seed bank. Genome Biol 14:R55. https://doi.org/10.1186/gb-2013-14-6-r55
Mantovani P, Maccaferri M, Sanguineti MC, Tuberosa R, Catizone I, Wenzl P et al (2008) An integrated DArT-SSR linkage map of durum wheat. Mol Breed 22:629–648. https://doi.org/10.1007/s11032-008-9205-3
Lam HM, Xu X, Liu X, Chen WB, Yang GH, Wong FL et al (2010) Resequencing of 31 wild and cultivated soybean genomes identifies patterns of genetic diversity and selection. Nat Genet 42:1053–1059. https://doi.org/10.1038/ng.715
Uitdewilligen JGML, Wolters AMA, D’hoop BB, Borm TJA, Visser RGF, van Eck HJ (2013) A next-generation sequencing method for genotyping-by-sequencing of highly heterozygous autotetraploid potato. PLoS One 8:e62355. https://doi.org/10.1371/journal.pone.0062355
Fu YB, Peterson GW (2011) Genetic diversity analysis with 454 pyrosequencing and genomic reduction confirmed the eastern and western division in the cultivated barley gene pool. Plant Genome 4:226–237
Lu F, Lipka AE, Glaubitz J, Elshire R, Cherney JH, Casler MD et al (2013) Switchgrass genomic diversity, ploidy, and evolution: novel insights from a network-based SNP discovery protocol. PLoS Genet 9(1):e1003215
Spindel J, Wright M, Chen C, Cobb J, Gage J, Harrington S et al (2013) Bridging the genotyping gap: using genotyping by sequencing (GBS) to add high-density SNP markers and new value to traditional bi-parental mapping and breeding populations. Theor Appl Genet 126:2699–2716. https://doi.org/10.1007/s00122-013-2166-x
Slavov GT, Nipper R, Robson P, Farrar K, Allison GG, Bosch M, Clifton-Brown JC et al (2014) Genome-wide association studies and prediction of 17 traits related to phenology, biomass and cell wall composition in the energy grass Miscanthus sinensis. New Phytol 201:1227–1239
Somegowda VK, Rayaprolu L, Rathore A, Deshpande SP, Gupta R (2021) Genome-wide association studies (GWAS) for traits related to fodder quality and biofuel in sorghum: progress and prospects. Protein Pept Lett 28:1. https://doi.org/10.2174/0929866528666210127153103
Young ND (1999) A cautiously optimistic vision for marker-assisted breeding. Mol Breed 5(6):505–510
Dukić M, Berner D, Roesti M, Haag CR, Ebert D (2016) A high-density genetic map reveals variation in recombination rate across the genome of Daphnia magna. BMC Genet 17:137. https://doi.org/10.1186/s12863-016-0445-7
Author information
Authors and Affiliations
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Rayaprolu, L., Deshpande, S.P., Gupta, R. (2022). Genotyping-by-Sequencing (GBS ) Method for Accelerating Marker-Assisted Selection (MAS) Program. In: Wani, S.H., Kumar, A. (eds) Genomics of Cereal Crops. Springer Protocols Handbooks. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-2533-0_12
Download citation
DOI: https://doi.org/10.1007/978-1-0716-2533-0_12
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2532-3
Online ISBN: 978-1-0716-2533-0
eBook Packages: Springer Protocols