Abstract
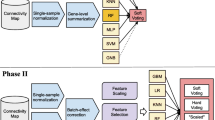
Machine learning (ML) already accelerates discoveries in many scientific fields and is the driver behind several new products. Recently, growing sample sizes enabled the use of ML approaches in larger omics studies. This work provides a guide through a typical analysis of an omics dataset using ML. As an example, this chapter demonstrates how to build a model predicting Drug-Induced Liver Injury based on transcriptomics data contained in the LINCS L1000 dataset. Each section covers best practices and pitfalls starting from data exploration and model training including hyperparameter search to validation and analysis of the final model. The code to reproduce the results is available at https://github.com/Evotec-Bioinformatics/ml-from-omics.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Subramanian A, Narayan R, Corsello SM et al (2017) A next generation connectivity map: L1000 platform and the first 1,000,000 profiles. Cell 171:1437–1452.e17
Liu Z, Thakkar S (2020) Deep learning on high-throughput transcriptomics to predict drug-induced liver injury. Front Bioeng Biotechnol 8:14
Walker PA, Ryder S, Lavado A et al (2020) The evolution of strategies to minimise the risk of human drug-induced liver injury (DILI) in drug discovery and development. Arch Toxicol 94:2559–2585
Leek J, Scharpf R, Bravo H et al (2010) Tackling the widespread and critical impact of batch effects in high-throughput data. Nat Rev Genet 11:733–739
McInnes L and Healy J (2018) UMAP: uniform manifold approximation and projection for dimension reduction. ArXiv abs/1802.03426
Narayan A, Berger B, Cho H (2020) Density-preserving data visualization unveils dynamic patterns of single-cell transcriptomic variability. bioRxiv
Van Rossum G, Drake FL (2009) Python 3 reference manual. CreateSpace, Scotts Valley, CA
Pedregosa F, Varoquaux G, Gramfort A et al (2011) Scikit-learn: machine learning in python. J Mach Learn Res 12:2825–2830
Li L, Jamieson K, DeSalvo G et al (2017) Hyperband: a novel bandit-based approach to Hyperparameter optimization. J Mach Learn Res 18:185:1–185:52
Falkner S, Klein A, Hutter F (2018) BOHB: robust and efficient hyperparameter optimization at scale. In: Dy J, Krause A (eds) Proceedings of the 35th International Conference on Machine Learning. PMLR, Stockholmsmässan, Stockholm Sweden, pp 1437–1446
Chicco D, Jurman G (2020) The advantages of the Matthews correlation coefficient (MCC) over F1 score and accuracy in binary classification evaluation. BMC Genomics 21:6
Institute of Medicine (2012) Evolution of translational omics: lessons learned and the path forward. The National Academies Press, Washington, DC
Carbon S, Douglass E, Good BM et al (2021) The gene ontology resource: enriching a GOld mine. Nucleic Acids Res 49:D325–D334
Acknowledgments
I am deeply grateful to my wife Sophia Rex and others for proofreading the final version of the manuscript. Additionally, I acknowledge the indispensable support of my parents Carmen and Michael Rex during the ongoing pandemic. Furthermore, I thank Thomas Siegmund, who is my superior at Evotec, for his constant encouragement and for giving me the opportunity to create this work.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2022 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Rex, R. (2022). Machine Learning from Omics Data . In: Heifetz, A. (eds) Artificial Intelligence in Drug Design. Methods in Molecular Biology, vol 2390. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1787-8_18
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1787-8_18
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1786-1
Online ISBN: 978-1-0716-1787-8
eBook Packages: Springer Protocols