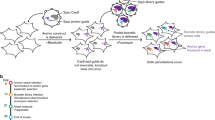
Abstract
Over the past two decades, the concept of synthetic lethality (SL) that queries genetic relationships between gene pairs has gradually emerged as one of the best strategies to selectively eliminate cancer cells. Some of the most successful approaches to identify synthetic lethal interactions (SLIs) were largely dependent on pooled screening formats that require heavy validation in order to mitigate false positives. Here, we describe a high-throughput method to identify SLIs using CRISPR-based strategy that covers, high-throughput production of plasmid DNA preparations, lentiviral production, and subsequent cellular transduction using single guide RNAs (sgRNAs). This method could be adopted to query hundreds of SLIs. As an example, we describe the methods associated with building an interaction map for DNA damage and repair (DDR) genes. The use of multiwell plates and image-based quantification allows a comparative measurement of SLIs at a high-resolution on a one-by-one basis. Furthermore, this scalable, arrayed CRISPR screening method can be applied to multiple cancer cell types, and genes of interest, resulting in new functional discoveries that can be exploited therapeutically.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Parameswaran S, Kundapur D, Vizeacoumar FS et al (2019) A road map to personalizing targeted cancer therapies using synthetic lethality. Trends Cancer 5(1):11–29. https://doi.org/10.1016/j.trecan.2018.11.001
Cunningham CE, MacAuley MJ, Yadav G et al (2019) Targeting the CINful genome: strategies to overcome tumor heterogeneity. Prog Biophys Mol Biol 147:77–91. https://doi.org/10.1016/j.pbiomolbio.2019.02.006
Paul JM, Templeton SD, Baharani A et al (2014) Building high-resolution synthetic lethal networks: a “Google map” of the cancer cell. Trends Mol Med 20(12):704–715. https://doi.org/10.1016/j.molmed.2014.09.009
Zhan T, Rindtorff N, Betge J et al (2018) CRISPR/Cas9 for cancer research and therapy. Semin Cancer Biol 55:106–119. https://doi.org/10.1016/j.semcancer.2018.04.001
Boone C, Bussey H, Andrews BJ (2007) Exploring genetic interactions and networks with yeast. Nat Rev Genet 8(6):437–449. https://doi.org/10.1038/nrg2085
Hengel SR, Spies MA, Spies M et al (2017) Small molecule inhibitors targeting DNA repair and DNA repair deficiency in research and cancer therapy. Cell Chem Biol 24(9):1101–1119. https://doi.org/10.1016/j.chembiol.2017.08.027.Small
Tsherniak A, Vazquez F, Montgomery PG et al (2017) Defining a cancer dependency map. Cell 170(3):564–576. https://doi.org/10.1016/j.cell.2017.06.010
Shen JP, Ideker T (2018) Synthetic lethal networks for precision oncology: promises and pitfalls. J Mol Biol 430(18 Pt A):2900–2912. https://doi.org/10.1016/j.jmb.2018.06.026
Cong L, Ran FA, Cox D et al (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. https://doi.org/10.1126/science.1231143.Multiplex
Wang T, Wei JJ, Sabatini DM et al (2012) Genetic screens in human cells using the CRISPR-Cas9 system. BMJ Support Palliat Care 2(3):256–263. https://doi.org/10.1136/bmjspcare-2011-000063
Bassik MC, Han K, Jeng EE et al (2017) Synergistic drug combinations for cancer identified in a CRISPR screen for pairwise genetic interactions. Nat Biotechnol 35(5):463–474. https://doi.org/10.1038/nbt.3834
Shalem O, Sanjana NE, Hartenian E et al (2014) Genome-scale CRISPR-Cas9 knockout screening in human cells. SSRN Electron J 343(6166):84–87. https://doi.org/10.4324/9781315853178
Doudna JA, Charpentier E (2014) The new frontier of genome engineering with CRISPR-Cas9. Science 346(6213). https://doi.org/10.1126/science.1258096
Davis AJ, Chen DJ (2013) DNA double strand break repair via non-homologous end-joining. Transl Cancer Res 2(3):130–143. https://doi.org/10.3978/j.issn.2218-676X.2013.04.02
Li X, Heyer WD (2008) Homologous recombination in DNA repair and DNA damage tolerance. Cell Res 18:99–113. https://doi.org/10.1038/cr.2008.1
Hart T, Tong AHY, Chan K et al (2017) Evaluation and design of genome-wide CRISPR/SpCas9 knockout screens. G3 (Bethesda) 7(8):2719–2727. https://doi.org/10.1534/g3.117.041277
Doench JG, Fusi N, Sullender M et al (2016) Optimized sgRNA design to maximize activity and minimize off-target effects of CRISPR-Cas9. Nat Biotechnol 34(2):184–191. https://doi.org/10.1038/nbt.3437
Collins SR, Roguev A, Krogan NJ (2010) Quantitative genetic interaction mapping using the E-MAP approach. Methods Enzymol 470:205–231. https://doi.org/10.1016/S0076-6879(10)70009-4
Krogan NJ, Cagney G, Yu H et al (2006) Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature 440(7084):637–643. https://doi.org/10.1038/nature04670
Tong AHY, Lesage G, Bader GD et al (2004) Global mapping of the yeast genetic interaction network. Science 303(5659):808–813. https://doi.org/10.1126/science.1091317
Metzakopian E, Strong A, Iyer V et al (2017) Enhancing the genome editing toolbox: genome wide CRISPR arrayed libraries. Sci Rep 7(1):1–9. https://doi.org/10.1038/s41598-017-01766-5
Acknowledgments
We thank members of the Vizeacoumar and Freywald laboratories for their feedback on this chapter. This work is supported by funding support from College of Medicine, University of Saskatchewan to M.J.M. and F.S.V.
Author contribution: F.S.V. and M.J.M. conceived and planned the strategies discussed in the chapter. M.J.M., O.A., and F.S.V. wrote or contributed to the writing of the manuscript.
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
MacAuley, M.J., Abuhussein, O., Vizeacoumar, F.S. (2021). Identification of Synthetic Lethal Interactions Using High-Throughput, Arrayed CRISPR/Cas9-Based Platforms. In: Vizeacoumar, F.J., Freywald, A. (eds) Mapping Genetic Interactions. Methods in Molecular Biology, vol 2381. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-1740-3_7
Download citation
DOI: https://doi.org/10.1007/978-1-0716-1740-3_7
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-1739-7
Online ISBN: 978-1-0716-1740-3
eBook Packages: Springer Protocols