Abstract
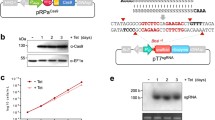
Chagas disease is a vector-borne tropical disease affecting millions of people worldwide, for which there is no vaccine or satisfactory treatment available. It is caused by the protozoan parasite Trypanosoma cruzi and considered endemic from North to South America. This parasite has unique metabolic and structural characteristics that make it an attractive organism for basic research. The genetic manipulation of T. cruzi has been historically challenging, as compared to other pathogenic protozoans. However, the use of the prokaryotic CRISPR/Cas9 system for genome editing has significantly improved the ability to generate genetically modified T. cruzi cell lines, becoming a powerful tool for the functional study of proteins in different stages of this parasite’s life cycle, including infective trypomastigotes and intracellular amastigotes. Using the CRISPR/Cas9 method that we adapted to T. cruzi, it has been possible to perform knockout, complementation and in situ tagging of T. cruzi genes. In our system we cotransfect T. cruzi epimastigotes with an expression vector containing the Cas9 sequence and a single guide RNA, together with a donor DNA template to promote DNA break repair by homologous recombination. As a result, we have obtained homogeneous populations of mutant epimastigotes using a single resistance marker to modify both alleles of the gene. Mitochondrial Ca2+ transport in trypanosomes is critical for shaping the dynamics of cytosolic Ca2+ increases, for the bioenergetics of the cells, and for viability and infectivity. In this chapter we describe the most effective methods to achieve genome editing in T. cruzi using as example the generation of mutant cell lines to study proteins involved in calcium homeostasis. Specifically, we describe the methods we have used for the study of three proteins involved in the calcium signaling cascade of T. cruzi: the inositol 1,4,5-trisphosphate receptor (TcIP3R), the mitochondrial calcium uniporter (TcMCU) and the calcium-sensitive pyruvate dehydrogenase phosphatase (TcPDP), using CRISPR/Cas9 technology as an approach to establish their role in the regulation of energy metabolism.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Horvath P, Barrangou R (2010) CRISPR/Cas, the immune system of bacteria and archaea. Science 327(5962):167–170. https://doi.org/10.1126/science.1179555
Rath D, Amlinger L, Rath A, Lundgren M (2015) The CRISPR-Cas immune system: biology, mechanisms and applications. Biochimie 117:119–128. https://doi.org/10.1016/j.biochi.2015.03.025
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, Hsu PD, Wu X, Jiang W, Marraffini LA, Zhang F (2013) Multiplex genome engineering using CRISPR/Cas systems. Science 339(6121):819–823. https://doi.org/10.1126/science.1231143
DiCarlo JE, Norville JE, Mali P, Rios X, Aach J, Church GM (2013) Genome engineering in Saccharomyces cerevisiae using CRISPR-Cas systems. Nucleic Acids Res 41(7):4336–4343. https://doi.org/10.1093/nar/gkt135
Jiang W, Zhou H, Bi H, Fromm M, Yang B, Weeks DP (2013) Demonstration of CRISPR/Cas9/sgRNA-mediated targeted gene modification in Arabidopsis, tobacco, sorghum and rice. Nucleic Acids Res 41(20):e188. https://doi.org/10.1093/nar/gkt780
Gratz SJ, Cummings AM, Nguyen JN, Hamm DC, Donohue LK, Harrison MM, Wildonger J, O’Connor-Giles KM (2013) Genome engineering of Drosophila with the CRISPR RNA-guided Cas9 nuclease. Genetics 194(4):1029–1035. https://doi.org/10.1534/genetics.113.152710
Ghorbal M, Gorman M, Macpherson CR, Martins RM, Scherf A, Lopez-Rubio JJ (2014) Genome editing in the human malaria parasite Plasmodium falciparum using the CRISPR-Cas9 system. Nat Biotechnol 32(8):819–821. https://doi.org/10.1038/nbt.2925
Lander N, Li ZH, Niyogi S, Docampo R (2015) CRISPR/Cas9-induced disruption of paraflagellar rod protein 1 and 2 genes in Trypanosoma cruzi reveals their role in flagellar attachment. MBio 6(4):e01012–e01015. https://doi.org/10.1128/mBio.01012-15
Sidik SM, Hackett CG, Tran F, Westwood NJ, Lourido S (2014) Efficient genome engineering of Toxoplasma gondii using CRISPR/Cas9. PLoS One 9(6):e100450. https://doi.org/10.1371/journal.pone.0100450
Peng D, Kurup SP, Yao PY, Minning TA, Tarleton RL (2015) CRISPR-Cas9-mediated single-gene and gene family disruption in Trypanosoma cruzi. MBio 6(1):e02097–e02014. https://doi.org/10.1128/mBio.02097-14
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science 337(6096):816–821. https://doi.org/10.1126/science.1225829
Sollelis L, Ghorbal M, MacPherson CR, Martins RM, Kuk N, Crobu L, Bastien P, Scherf A, Lopez-Rubio JJ, Sterkers Y (2015) First efficient CRISPR-Cas9-mediated genome editing in Leishmania parasites. Cell Microbiol 17(10):1405–1412. https://doi.org/10.1111/cmi.12456
Lander N, Chiurillo MA, Docampo R (2016) Genome editing by CRISPR/Cas9: a game change in the genetic manipulation of protists. J Eukaryot Microbiol 63(5):679–690. https://doi.org/10.1111/jeu.12338
Forsyth CJ, Hernandez S, Olmedo W, Abuhamidah A, Traina MI, Sanchez DR, Soverow J, Meymandi SK (2016) Safety profile of nifurtimox for treatment of Chagas disease in the United States. Clin Infect Dis 63(8):1056–1062. https://doi.org/10.1093/cid/ciw477
Docampo R (2011) Molecular parasitology in the 21st century. Essays Biochem 51:1–13. https://doi.org/10.1042/bse0510001
Lander N, Chiurillo MA, Storey M, Vercesi AE, Docampo R (2016) CRISPR/Cas9-mediated endogenous C-terminal tagging of Trypanosoma cruzi genes reveals the acidocalcisome localization of the inositol 1,4,5-trisphosphate receptor. J Biol Chem 291(49):25505–25515. https://doi.org/10.1074/jbc.M116.749655
Chiurillo MA, Lander N, Bertolini MS, Storey M, Vercesi AE, Docampo R (2017) Different roles of mitochondrial calcium uniporter complex subunits in growth and infectivity of Trypanosoma cruzi. MBio 8(3):e00574–e00517. https://doi.org/10.1128/mBio.00574-17
Soares Medeiros LC, South L, Peng D, Bustamante JM, Wang W, Bunkofske M, Perumal N, Sanchez-Valdez F, Tarleton RL (2017) Rapid, selection-free, high-efficiency genome editing in protozoan parasites using CRISPR-Cas9 ribonucleoproteins. MBio 8(6):e01788–e01717. https://doi.org/10.1128/mBio.01788-17
Costa FC, Francisco AF, Jayawardhana S, Calderano SG, Lewis MD, Olmo F, Beneke T, Gluenz E, Sunter J, Dean S, Kelly JM, Taylor MC (2018) Expanding the toolbox for Trypanosoma cruzi: a parasite line incorporating a bioluminescence-fluorescence dual reporter and streamlined CRISPR/Cas9 functionality for rapid in vivo localisation and phenotyping. PLoS Negl Trop Dis 12(4):e0006388. https://doi.org/10.1371/journal.pntd.0006388
Lander N, Chiurillo MA, Bertolini MS, Storey M, Vercesi AE, Docampo R (2018) Calcium-sensitive pyruvate dehydrogenase phosphatase is required for energy metabolism, growth, differentiation, and infectivity of Trypanosoma cruzi. J Biol Chem 293(45):17402–17417. https://doi.org/10.1074/jbc.RA118.004498
Lander N, Chiurillo MA, Vercesi AE, Docampo R (2017) Endogenous C-terminal tagging by CRISPR/Cas9 in Trypanosoma cruzi. Bio Protoc 7(10). https://doi.org/10.21769/BioProtoc.2299
Moreno SN, Silva J, Vercesi AE, Docampo R (1994) Cytosolic-free calcium elevation in Trypanosoma cruzi is required for cell invasion. J Exp Med 180(4):1535–1540
Lammel EM, Barbieri MA, Wilkowsky SE, Bertini F, Isola EL (1996) Trypanosoma cruzi: involvement of intracellular calcium in multiplication and differentiation. Exp Parasitol 83(2):240–249. https://doi.org/10.1006/expr.1996.0070
Rohloff P, Rodrigues CO, Docampo R (2003) Regulatory volume decrease in Trypanosoma cruzi involves amino acid efflux and changes in intracellular calcium. Mol Biochem Parasitol 126(2):219–230
Engman DM, Krause KH, Blumin JH, Kim KS, Kirchhoff LV, Donelson JE (1989) A novel flagellar Ca2+-binding protein in trypanosomes. J Biol Chem 264(31):18627–18631
Huang G, Vercesi AE, Docampo R (2013) Essential regulation of cell bioenergetics in Trypanosoma brucei by the mitochondrial calcium uniporter. Nat Commun 4:2865. https://doi.org/10.1038/ncomms3865
Docampo R, Vercesi AE (1989) Ca2+ transport by coupled Trypanosoma cruzi mitochondria in situ. J Biol Chem 264(1):108–111
Vercesi AE, Bernardes CF, Hoffmann ME, Gadelha FR, Docampo R (1991) Digitonin permeabilization does not affect mitochondrial function and allows the determination of the mitochondrial membrane potential of Trypanosoma cruzi in situ. J Biol Chem 266(22):14431–14434
Oberholzer M, Morand S, Kunz S, Seebeck T (2006) A vector series for rapid PCR-mediated C-terminal in situ tagging of Trypanosoma brucei genes. Mol Biochem Parasitol 145(1):117–120. https://doi.org/10.1016/j.molbiopara.2005.09.002
Bone GJ, Steinert M (1956) Isotopes incorporated in the nucleic acids of Trypanosoma mega. Nature 178:2. https://doi.org/10.1038/178308a0
Aslett M, Aurrecoechea C, Berriman M, Brestelli J, Brunk BP, Carrington M, Depledge DP, Fischer S, Gajria B, Gao X, Gardner MJ, Gingle A, Grant G, Harb OS, Heiges M, Hertz-Fowler C, Houston R, Innamorato F, Iodice J, Kissinger JC, Kraemer E, Li W, Logan FJ, Miller JA, Mitra S, Myler PJ, Nayak V, Pennington C, Phan I, Pinney DF, Ramasamy G, Rogers MB, Roos DS, Ross C, Sivam D, Smith DF, Srinivasamoorthy G, Stoeckert CJ Jr, Subramanian S, Thibodeau R, Tivey A, Treatman C, Velarde G, Wang H (2010) TriTrypDB: a functional genomic resource for the Trypanosomatidae. Nucleic Acids Res 38(Database issue):D457–D462. https://doi.org/10.1093/nar/gkp851
Peng D, Tarleton R (2015) EuPaGDT: a web tool tailored to design CRISPR guide RNAs for eukaryotic pathogens. Microb Genom 1(4):e000033. https://doi.org/10.1099/mgen.0.000033
Zingales B, Andrade SG, Briones MR, Campbell DA, Chiari E, Fernandes O, Guhl F, Lages-Silva E, Macedo AM, Machado CR, Miles MA, Romanha AJ, Sturm NR, Tibayrenc M, Schijman AG, Second Satellite M (2009) A new consensus for Trypanosoma cruzi intraspecific nomenclature: second revision meeting recommends TcI to TcVI. Mem Inst Oswaldo Cruz 104(7):1051–1054
Acknowledgments
We thank Prof. Anibal E. Vercesi (University of Campinas, Brazil) for his encouragement and advice. This work was funded by the U.S. National Institutes of Health (grants AI107663 and AI140421).
Author information
Authors and Affiliations
Corresponding authors
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Lander, N., Chiurillo, M.A., Docampo, R. (2020). CRISPR/Cas9 Technology Applied to the Study of Proteins Involved in Calcium Signaling in Trypanosoma cruzi. In: Michels, P., Ginger, M., Zilberstein, D. (eds) Trypanosomatids. Methods in Molecular Biology, vol 2116. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0294-2_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0294-2_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0293-5
Online ISBN: 978-1-0716-0294-2
eBook Packages: Springer Protocols