Abstract
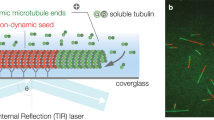
Microtubules are dynamic non-covalent mesoscopic polymers. Their dynamic behavior is essential for cell biological processes ranging from intracellular transport to cell division and neurogenesis. Fluorescence microscopy has been the method of choice for monitoring microtubule dynamics in the last two decades. However, fluorescent microtubules are prone to photodamage that alters their dynamics, and the fluorescent label itself can affect microtubule properties. Dark-field imaging is a label-free technique that can generate high signal-to-noise, low-background images of microtubules at high acquisition rates without the photobleaching inherent to fluorescence microscopy. Here, we describe how to image in vitro microtubule dynamics using dark-field microscopy. The ability to image microtubules label-free allows the investigation of the dynamic properties of non-abundant tubulin species where fluorescent labeling is not feasible, free from the confounding effects arising from the addition of fluorescent labels.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Kuriyama R, Miki-Noumura T (1975) Light-microscopic observations of individual microtubules reconstituted from brain tubulin. J Cell Sci 19(3):607–620
Mitchison T, Kirschner M (1984) Dynamic instability of microtubule growth. Nature 312(5991):237
Walker RA, O’Brien ET, Pryer NK, Soboeiro MF, Voter WA, Erickson HP, Salmon ED (1988) Dynamic instability of individual microtubules analyzed by video light microscopy: rate constants and transition frequencies. J Cell Biol 107(4):1437–1448
Horio T, Hotani H (1986) Visualization of the dynamic instability of individual microtubules by dark-field microscopy. Nature 321(6070):605
Summers K, Kirschner MW (1979) Characteristics of the polar assembly and disassembly of microtubules observed in vitro by darkfield light microscopy. J Cell Biol 83(1):205–217
Akhmanova A, Steinmetz MO (2015) Control of microtubule organization and dynamics: two ends in the limelight. Nat Rev Mol Cell Biol 16(12):711–726. https://doi.org/10.1038/nrm4084
Guo H, Xu C, Liu C, Qu E, Yuan M, Li Z, Cheng B, Zhang D (2006) Mechanism and dynamics of breakage of fluorescent microtubules. Biophys J 90(6):2093–2098
Aumeier C, Schaedel L, Gaillard J, John K, Blanchoin L, Thery M (2016) Self-repair promotes microtubule rescue. Nat Cell Biol 18(10):1054–1064. https://doi.org/10.1038/ncb3406
Vigers GP, Coue M, McIntosh JR (1988) Fluorescent microtubules break up under illumination. J Cell Biol 107(3):1011–1024
Widlund PO, Podolski M, Reber S, Alper J, Storch M, Hyman AA, Howard J, Drechsel DN (2012) One-step purification of assembly-competent tubulin from diverse eukaryotic sources. Mol Biol Cell 23(22):4393–4401. https://doi.org/10.1091/mbc.E12-06-0444
Vemu A, Garnham CP, Lee DY, Roll-Mecak A (2014) Generation of differentially modified microtubules using in vitro enzymatic approaches. Methods Enzymol 540:149–166. https://doi.org/10.1016/B978-0-12-397924-7.00009-1
Vemu A, Atherton J, Spector JO, Szyk A, Moores CA, Roll-Mecak A (2016) Structure and dynamics of single-isoform recombinant neuronal human tubulin. J Biol Chem 291(25):12907–12915. https://doi.org/10.1074/jbc.C116.731133
Minoura I, Hachikubo Y, Yamakita Y, Takazaki H, Ayukawa R, Uchimura S, Muto E (2013) Overexpression, purification, and functional analysis of recombinant human tubulin dimer. FEBS Lett 587(21):3450–3455. https://doi.org/10.1016/j.febslet.2013.08.032
Ti SC, Pamula MC, Howes SC, Duellberg C, Cade NI, Kleiner RE, Forth S, Surrey T, Nogales E, Kapoor TM (2016) Mutations in human tubulin proximal to the kinesin-binding site alter dynamic instability at microtubule plus- and minus-ends. Dev Cell 37(1):72–84. https://doi.org/10.1016/j.devcel.2016.03.003
Johnson V, Ayaz P, Huddleston P, Rice LM (2011) Design, overexpression, and purification of polymerization-blocked yeast alphabeta-tubulin mutants. Biochemistry 50(40):8636–8644. https://doi.org/10.1021/bi2005174
Chaaban S, Jariwala S, Hsu CT, Redemann S, Kollman JM, Muller-Reichert T, Sept D, Bui KH, Brouhard GJ (2018) The structure and dynamics of C. elegans tubulin reveals the mechanistic basis of microtubule growth. Dev Cell 47(2):191–204, e198. https://doi.org/10.1016/j.devcel.2018.08.023
Weisenberg RC (1972) Microtubule formation in vitro in solutions containing low calcium concentrations. Science 177(4054):1104–1105
Vemu A, Atherton J, Spector JO, Moores CA, Roll-Mecak A (2017) Tubulin isoform composition tunes microtubule dynamics. Mol Biol Cell 28(25):3564–3572. https://doi.org/10.1091/mbc.E17-02-0124
Geyer EA, Burns A, Lalonde BA, Ye X, Piedra FA, Huffaker TC, Rice LM (2015) A mutation uncouples the tubulin conformational and GTPase cycles, revealing allosteric control of microtubule dynamics. Elife 4:e10113. https://doi.org/10.7554/eLife.10113
Andrecka J, Arroyo JO, Lewis K, Cross RA, Kukura P (2016) Label-free imaging of microtubules with sub-nm precision using interferometric scattering microscopy. Biophys J 110(1):214–217
Mahamdeh M, Simmert S, Luchniak A, Schaeffer E, Howard J (2018) Label-free high-speed wide-field imaging of single microtubules using interference reflection microscopy. J Microsc 272(1):60–66
Simmert S, Abdosamadi MK, Hermsdorf G, Schäffer E (2018) LED-based interference-reflection microscopy combined with optical tweezers for quantitative three-dimensional microtubule imaging. Opt Express 26(11):14499–14513
Katsuki M, Drummond DR, Osei M, Cross RA (2009) Mal3 masks catastrophe events in Schizosaccharomyces pombe microtubules by inhibiting shrinkage and promoting rescue. J Biol Chem 284(43):29246–29250. https://doi.org/10.1074/jbc.C109.052159
Kandel ME, Teng KW, Selvin PR, Popescu G (2017) Label-free imaging of single microtubule dynamics using spatial light interference microscopy. ACS Nano 11(1):647–655. https://doi.org/10.1021/acsnano.6b06945
Ueno H, Nishikawa S, Iino R, Tabata KV, Sakakihara S, Yanagida T, Noji H (2010) Simple dark-field microscopy with nanometer spatial precision and microsecond temporal resolution. Biophys J 98(9):2014–2023. https://doi.org/10.1016/j.bpj.2010.01.011
Edelstein AD, Tsuchida MA, Amodaj N, Pinkard H, Vale RD, Stuurman N (2014) Advanced methods of microscope control using muManager software. J Biol Methods 1(2):e10. https://doi.org/10.14440/jbm.2014.36
Ziółkowska NE, Roll-Mecak A (2013) In vitro microtubule severing assays. Methods Mol Biol 1046:323–334. https://doi.org/10.1007/978-1-62703-538-5_19; pmid: 23868597
Schindelin J, Arganda-Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open-source platform for biological-image analysis. Nat Methods 9(7):676–682. https://doi.org/10.1038/nmeth.2019
Mukherjee A, Jenkins B, Fang C, Radke RJ, Banker G, Roysam B (2011) Automated kymograph analysis for profiling axonal transport of secretory granules. Med Image Anal 15(3):354–367. https://doi.org/10.1016/j.media.2010.12.005
Kapoor V, Hirst WG, Hentschel C, Preibisch S, Reber S (2019) MTrack: automated detection, tracking, and analysis of dynamic microtubules. Sci Rep 9(1):3794. https://doi.org/10.1038/s41598-018-37767-1
Ruhnow F, Zwicker D, Diez S (2011) Tracking single particles and elongated filaments with nanometer precision. Biophys J 100(11):2820–2828. https://doi.org/10.1016/j.bpj.2011.04.023
Acknowledgment
A.R.M. is supported by the intramural programs of the National Institute of Neurological Disorders and Stroke (NINDS) and the National, Heart, Lung, and Blood Institute (NHLBI).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
Spector, J.O., Vemu, A., Roll-Mecak, A. (2020). In Vitro Microtubule Dynamics Assays Using Dark-Field Microscopy. In: Maiato, H. (eds) Cytoskeleton Dynamics. Methods in Molecular Biology, vol 2101. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0219-5_4
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0219-5_4
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0218-8
Online ISBN: 978-1-0716-0219-5
eBook Packages: Springer Protocols