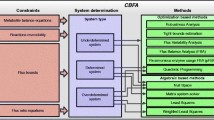
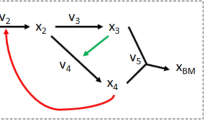
Abstract
As genetic engineering of organisms has grown easier and more precise, computational modeling of metabolic systems has played an increasingly important role in both guiding experimental interventions and in understanding the results of metabolic perturbations.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Orth JD, Thiele I, Palsson BØ (2010) What is flux balance analysis? Nat Biotechnol 28:245–248. https://doi.org/10.1038/nbt.1614
Lewis NE, Nagarajan H, Palsson BO (2012) Constraining the metabolic genotypephenotype relationship using a phylogeny of in silico methods. Nat Rev Microbiol. https://doi.org/10.1038/nrmicro2737
Feist AM, Palsson BO (2010) The biomass objective function. Curr Opin Microbiol 13:344–349. https://doi.org/10.1016/j.mib.2010.03.003
Spielman DA, Teng S-H (2004) Smoothed analysis of algorithms. J ACM 51:385–463. https://doi.org/10.1145/990308.990310
Thiele I, Palsson BØ (2010) A protocol for generating a high-quality genome-scale metabolic reconstruction. Nat Protoc 5:93–121. https://doi.org/10.1038/nprot.2009.203
Burgard AP, Pharkya P, Maranas CD (2003) Optknock: a bilevel programming framework for identifying gene knockout strategies for microbial strain optimization. Biotechnol Bioeng 84:647–657. https://doi.org/10.1002/bit.10803
Mahadevan R, Schilling C (2003) The effects of alternate optimal solutions in constraint-based genome-scale metabolic models. Metab Eng 5:264–276. https://doi.org/10.1016/j.ymben.2003.09.002
Segre D, Vitkup D, Church GM (2002) Analysis of optimality in natural and perturbed metabolic networks. Proc Natl Acad Sci U S A 99:15112–15117. https://doi.org/10.1073/pnas.232349399
Shlomi T, Berkman O, Ruppin E (2005) Regulatory on/off minimization of metabolic flux changes after genetic perturbations. Proc Natl Acad Sci U S A 102:7695–7700. https://doi.org/10.1073/pnas.0406346102
Fong SS, Burgard AP, Herring CD et al (2005) In silico design and adaptive evolution of Escherichia coli for production of lactic acid. Biotechnol Bioeng 91:643–648. https://doi.org/10.1002/bit.20542
Zanghellini J, Ruckerbauer DE, Hanscho M, Jungreuthmayer C (2013) Elementary flux modes in a nutshell: properties, calculation and applications. Biotechnol J 8:1009–1016. https://doi.org/10.1002/biot.201200269
Hädicke O, Klamt S (2011) Computing complex metabolic intervention strategies using constrained minimal cut sets. Metab Eng 13:204–213. https://doi.org/10.1016/j.ymben.2010.12.004
Shen CR, Lan EI, Dekishima Y et al (2011) Driving forces enable high-titer anaerobic 1-butanol synthesis in Escherichia coli. Appl Environ Microbiol 77:2905–2915. https://doi.org/10.1128/aem.03034-10
Machado D, Herrgård MJ (2015) Co-evolution of strain design methods based on flux balance and elementary mode analysis. Metab Eng Commun 2:85–92. https://doi.org/10.1016/j.meteno.2015.04.001
King ZA, Lu J, Dräger A et al (2015) BiGG models: a platform for integrating, standardizing and sharing genome-scale models. Nucleic Acids Res 44:D515–D522. https://doi.org/10.1093/nar/gkv1049
Henry CS, DeJongh M, Best AA et al (2010) High-throughput generation, optimization and analysis of genome-scale metabolic models. Nat Biotechnol 28:977–982. https://doi.org/10.1038/nbt.1672
Klamt S, Saez-Rodriguez J, Gilles ED (2007) BMC Syst Biol 1:2. https://doi.org/10.1186/1752-0509-1-2
Agren R, Liu L, Shoaie S et al (2013) The RAVEN toolbox and its use for generating a genome-scale metabolic model for penicillium chrysogenum. PLoS Comput Biol 9:e1002980. https://doi.org/10.1371/journal.pcbi.1002980
Hyduke D, Hyduke D, Schellenberger J et al (2011) COBRA toolbox 2.0. Protocol Exchange. https://doi.org/10.1038/protex.2011.234
Ebrahim A, Lerman JA, Palsson BO, Hyduke DR (2013) COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst Biol 7:74. https://doi.org/10.1186/1752-0509-7-74
Lewis NE, Hixson KK, Conrad TM et al (2010) Omic data from evolved E. coli are consistent with computed optimal growth from genome-scale models. Mol Syst Biol 6. https://doi.org/10.1038/msb.2010.47
Kanehisa M, Furumichi M, Tanabe M et al (2016) KEGG: new perspectives on genomes, pathways, diseases and drugs. Nucleic Acids Res 45:D353–D361. https://doi.org/10.1093/nar/gkw1092
Caspi R, Altman T, Billington R et al (2013) The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res 42:D459–D471. https://doi.org/10.1093/nar/gkt1103
McKinlay JB, Shachar-Hill Y, Zeikus JG, Vieille C (2007) Determining actinobacillus succinogenes metabolic pathways and fluxes by NMR and GC-MS analyses of 13C-labeled metabolic product isotopomers. Metab Eng 9:177–192. https://doi.org/10.1016/j.ymben.2006.10.006
Orth JD, Palsson BØ, Fleming RMT (2010) Reconstruction and use of microbial metabolic networks: the core Escherichia coli metabolic model as an educational guide. EcoSal Plus 4. https://doi.org/10.1128/ecosalplus.10.2.1
King ZA, Dräger A, Ebrahim A et al (2015) Escher: a web application for building, sharing, and embedding data-rich visualizations of biological pathways. PLoS Comput Biol 11:e1004321. https://doi.org/10.1371/journal.pcbi.1004321
Acknowledgments
This work was funded by the U.S. Department of Energy’s Bioenergy Technologies Office (DOE-BETO). This work was authored in part by Alliance for Sustainable Energy, LLC, the Manager and Operator of the National Renewable Energy Laboratory for the U.S. Department of Energy (DOE) under Contract No. DE-AC36-08GO28308. The views expressed in the chapter do not necessarily represent the views of the DOE or the U.S. Government. The U.S. Government retains, and the publisher, by accepting the article for publication, acknowledges that the U.S. Government retains a nonexclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for U.S. Government purposes.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Science+Business Media, LLC, part of Springer Nature
About this protocol
Cite this protocol
St. John, P.C., Bomble, Y.J. (2020). Software and Methods for Computational Flux Balance Analysis. In: Himmel, M., Bomble, Y. (eds) Metabolic Pathway Engineering. Methods in Molecular Biology, vol 2096. Humana, New York, NY. https://doi.org/10.1007/978-1-0716-0195-2_13
Download citation
DOI: https://doi.org/10.1007/978-1-0716-0195-2_13
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-0194-5
Online ISBN: 978-1-0716-0195-2
eBook Packages: Springer Protocols