Abstract
SLC25A42 is an inner mitochondrial membrane protein which has been shown to transport coenzyme A through a lipid bilayer in vitro. A homozygous missense variant in this gene has been recently reported in 13 subjects of Arab descent presenting with mitochondriopathy with variable clinical manifestations. By exome sequencing, we identified two additional individuals carrying rare variants in this gene. One subject was found to carry the previously reported missense variant in homozygous state, while the second subject carried a homozygous canonical splice site variant resulting in a splice defect. With the identification of two additional cases, we corroborate the association between rare variants in SLC25A42 and a clinical presentation characterized by myopathy, developmental delay, lactic acidosis, and encephalopathy. Furthermore, we highlight the biochemical consequences of the splice defect by measuring a mild decrease of coenzyme A content in SLC25A42-mutant fibroblasts.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Introduction
The mitochondrial matrix hosts a variety of vital enzymatic functions including fatty acid and pyruvate oxidation and the citric acid cycle. As the matrix is surrounded by a double membrane, mitochondrial carriers are required for an efficient exchange with the cytosol of metabolites, nucleotides, and cofactors. To date 53 members of the solute carrier family 25 (SLC25) have been identified (Palmieri and Monne 2016; Palmieri 2004, 2013, 2014). In vitro experiments have suggested that SLC25A42 is able to transport adenine nucleotides, adenosine 3’, 5’-diphosphate (PAP), and to a lesser extent coenzyme A (CoA) and dephosphoCoA (dPCoA) through a phospholipid bilayer (Fiermonte et al. 2009). It has been proposed that SLC25A42 could import cytosolic CoA or dPCoA in mitochondria in counter exchange with PAP or adenine nucleotides (Fiermonte et al. 2009), allowing mitochondria and cytosol to keep their CoA pools separated (Leonardi et al. 2005). However, the identification of coenzyme A synthase (COASY) in the mitochondrial matrix converting 4-phosphopantetheine to CoA made this hypothesis questionable (Dusi et al. 2014). Based on the finding that CG4241, the Drosophila ortholog of SLC25A42, transports dPCoA into mitochondria (Vozza et al. 2017), it has been proposed that SLC25A42 might exchange mitochondrial dPCoA with cytosolic ADP, allowing CoA to be produced both in the matrix by COASY and in the cytosol by the monofunctional dPCoA kinase. CoA plays an essential role in various anabolic and catabolic pathways as well as regulation of cellular processes via allosteric interactions and gene expression. These include the biosynthesis of fatty acids, ketone bodies, and cholesterol, amino acid metabolism, fatty acid oxidation, biosynthesis neurotransmitter acetylcholine, and acetylation of histones and regulation of gene expression. Defects within three of the five enzymatic steps involved in CoA biosynthesis have been linked both to childhood-onset forms of neurodegeneration with brain iron accumulation (PANK2, COASY) (Dusi et al. 2014; Bosveld et al. 2008) and to dilated cardiomyopathy with no neurodegeneration (PPCS) (Iuso et al. 2018).
In 2016, Shamseldin et al. identified a missense mutation in a highly conserved amino acid of SLC25A42 in a subject presenting with mitochondrial myopathy characterized by muscle weakness, chronic fatigue, and dysarthria (Shamseldin et al. 2016). The same group reported that SLC25A42 knockdown in zebra fish caused morphological anomalies and motor defects which are rescued upon injection of the human wild-type SLC25A42 mRNA, but not the mRNA carrying the subject’s variant (Shamseldin et al. 2016). Recently, Almannai et al. identified the same founder mutation in 12 additional individuals presenting with variable manifestations ranging from asymptomatic lactic acidosis to a severe phenotype characterized by developmental regression and encephalopathy (Almannai et al. 2018). In the current manuscript, we report on the identification of a novel homozygous canonical splice site variant and the previously described missense homozygous variant in SLC25A42 in individuals with a metabolic encephalopathy thereby confirming the role of SLC25A42 mutations in human disease. Moreover, we provide tentative evidence that SLC25A42 loss of function (LOF) mutations can cause indeed decreased cellular coenzyme A.
Subjects and Methods
Subjects
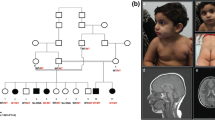
Case 1 is a 6-year-old Saudi boy born preterm at 34 weeks of gestation after a normal pregnancy. His consanguineous parents (second-degree relatives) as well as the two older sisters are healthy. After birth, the patient had delayed respiratory adaptation which required the admission in a neonatal intensive care unit and sepsis. He showed repetitive pulmonary infections during the first year of life and recurrent episodes of gastroesophageal reflux. He started crawling at 24 months of age, but currently he is not able to sit or walk. Diagnostic follow-up with electromyography, nerve conduction study, and metabolic investigations (VLCFA, organic acids, guanidino metabolites) did not reveal any specific diagnosis. MRI scan at the age of 1 year was normal, but at the age of 3 years showed symmetric hyperintensities in the putamen (Fig. 1). The boy developed dystonia, profound muscular hypotonia, and severe cognitive impairment with absent speech. His workup included elevated levels of lactic acid. Clinical features are summarized in Supplementary Table 1.
Case 2 is a 9-year-old boy born at term to healthy unrelated German parents with normal birth measurements (birth weight, 2,650 g; APGAR scores, 10/10; cord blood pH. not known). At the age of 1 year, he had an upper airway infection with fever and loss of appetite for 4 days and was admitted with a metabolic crisis at the fifth day in a comatose state. His metabolic workup showed rhabdomyolysis with elevated creatine kinase activity (12,000 U/L), hypoglycemia (30 mg/dL), hyperammonemia (300 μmol/L), and severe metabolic acidosis with elevated liver enzymes (GOT 346 U/L, GTP 78 U/L, LDH 540 U/L). Lactate levels were persistently elevated. Abdominal sonography showed an enlarged liver without structural changes, and an echocardiography was unremarkable. Brain CT scans were normal, but the EEG showed clear increase of delta waves. His clinical condition stabilized, but 1 month later during another febrile episode, he developed tonic-clonic seizure. These crises of fever and seizures repeatedly occurred over the years necessitating anti-epileptic medication. Bayley scale testing showed that his psychomotor development was delayed throughout the disease course. Specifically, the patient showed mild linguistic impairment, behavioral disorder (with impulsivity and aggressiveness), and mild generalized muscular hypotonia. Testing for very-long-chain acyl-CoA dehydrogenase deficiency (VLCAD) showed a slightly reduction of palmitoyl-CoA oxidation. Biochemical analysis of a muscle biopsy at the age of 2 years showed normal activities of respiratory chain complexes and PDHc, and histological examination was normal. Testing glycosylation patterns were unremarkable. At age of 3 years, his TSH level was slightly elevated (4.25 μU/mL), and he was treated with iodine. His EEG follow-up through his growth was unremarkable. MRI scan at age of 9 years showed abnormal bilateral signal alterations in the caudate nucleus (Fig. 1). Clinical features are summarized in Supplementary Table 1.
Genetic Studies
Exome sequencing and variant filtering were performed as described previously (Haack et al. 2010). Briefly, coding DNA fragments were enriched with the SureSelect Human All Exon 50 Mb V5 Kit (Agilent, Santa Clara, CA, USA), and sequencing was performed on a HiSeq2500 system (Illumina, San Diego, CA, USA). Reads were aligned to the human genome assembly hg19 (UCSC Genome Browser) with Burrows-Wheeler Aligner (version 0.7.5), and detection of genetic variation was performed using SAMtools (version 0.1.19), Pindel (version 0.2.5a7), and ExomeDepth (version 1.0.0). 97.6% (Case 1) and 97.9% (Case 2) of the target were covered at least 20-fold.
RNA Sequencing and RT-PCR
RNA sequencing was performed as described (Haack et al. 2013). RNA was isolated from whole cell lysates using the AllPrep RNA Kit (Qiagen), and RNA integrity number (RIN) was determined with the Agilent 2100 Bioanalyzer (RNA 6000 Nano Kit, Agilent). For library preparation, 1 μg of RNA was poly(A) selected, fragmented, and reverse transcribed with the Elute, Prime, Fragment Mix (Illumina). End repair, A-tailing, adaptor ligation, and library enrichment were performed as described in the low-throughput protocol of the TruSeq RNA Sample Prep Guide (Illumina). RNA libraries were assessed for quality and quantity with the Agilent 2100 Bioanalyzer and the Quant-iT PicoGreen dsDNA Assay Kit (Life Technologies). RNA libraries were sequenced as 100 bp paired-end runs on an Illumina HiSeq2500 platform.
For the RT-PCR, 1 μg of RNA was reverse transcribed with M-MLV reverse transcriptase (Promega, GmbH), and oligo dT. SLC25A42 (NM_178526.4) exons were amplified with Thermo-Start Taq DNA Polymerase (ABgene, Epsom, UK) and primers listed in Supplementary Fig. 1a, applying the conditions described in Supplementary Fig. 1b, c.
Measure of Mitochondrial Content of Coenzyme A
One million cells were collected and spun down, and the pellet was resuspended in PBS in the presence of 0.5% NP40. The cellular membranes were broken by passing the suspension through a 1 mL insulin syringe ten times. Unbroken cells were removed by centrifugation at 1,500 rpm for 5 min. The cleared supernatant was recovered and used for the CoA measure according the manufacturer instructions (Abcam, ab138889). The kit is based on an ultrasensitive (detection limit of 40 nM) fluorometric detection of –SH group in CoA. It contains a proprietary fluorogenic acetyl-CoA green indicator dye that becomes strongly fluorescent upon reacting with –SH. For each measure, 50 μL of cleared cellular supernatant was used. Citation 3 was used for measuring fluorescence (BioTek).
Results
Identification of Rare Variants in SLC25A42
Exome data detected about 290 Mb regions of homozygosity in Case 1 confirming the consanguinity within the family. Based on that, a search for rare homozygous variants with minor allele frequency (MAF) <0.1% in our in-house database comprising ~12,000 control exomes (04/2018) was performed. This search resulted in homozygous variants in 29 genes, which have been afterward filtered for previously reported genes in OMIM (phenotype key 3), HGMD, or ClinVar databases. This filtering step prioritized variants in three genes (SLC25A42, UPB1, and HEATR2) associated with autosomal recessive diseases. We considered SLC25A42, harboring the homozygous missense variant c.871A>G, p.Asn291Asp, as the best candidate gene based on the fact that the variant c.871A>G was previously reported as clinically relevant in individuals with overlapping clinical features (Almannai et al. 2018; Shamseldin et al. 2016). Furthermore, the variant is predicted to be damaging by in silico prediction programs (pph2, Sift, CADD). The variants in the other two genes (UPB1 and HEATR2) did not fit or explain the phenotype of Case 1.
In Case 2, assuming a recessive mode of inheritance, a search for homozygous, potentially compound heterozygous, or X-chromosomal non-synonymous rare variants (MAF <0.1%) in our in-house database prioritized only one LOF variant, the homozygous splice change c.380+2T>A, p.(?) in SLC25A42. This variant is predicted to affect canonical splice donor site of intron 5 (Fig. 2A). The variant is present only once in heterozygous state in GnomAD database.
SLC25A42 gene and transcripts. (A) Schematic representation of the SLC25A42 gene (NC_000019.10), and localization of the variant c. 380+2T>A in Case 2. The same length has been used for all introns. The mutation nomenclature is based on the splice variant NM_178526.4. (B) Sashimi plots summarizing the analysis of transcriptome data for SLC25A42 in Case 2 and two unrelated control fibroblasts (Controls 1 and 2). The canonical transcript of SLC25A42 is drawn at the top (NM_178526.4), while the three alternative transcripts are indicated in lowercase (a, b, c) directly on the Case 2 plot. (C) PCR products of cDNA from a control (C) and Case 2 (P) using primers spanning regions: exon 2–exon 7 (2–7), exon 3–exon 7 (3–7), and exon 6–exon 7 (6–7). M1 and M2 indicate 100 bp and 1 kb ladders, respectively. (D) Schematic representation of the canonical splicing (blue lines) producing the canonical transcript NM_178526.4, and the aberrant splicing (dashed red lines) producing alternative transcripts. Primer pairs used for the RT-PCR are indicated on the top of the canonical transcript
Sanger sequencing was used to confirm the identified mutations and test the carrier status of unaffected parents. Oligonucleotide sequences and PCR conditions are available upon request.
The c.380+2T>A Variant Abolishes the Canonical Splicing of SLC25A42
We investigated the possibility of alternative transcripts by analyzing RNA sequencing data generated from Case 2 and from control fibroblasts. Figure 2B summarizes in Sashimi plots all detected RNA splice variants. In control fibroblasts the only transcribed RNA was the canonical SLC25A42 transcript (NM_178526.4). In Case 2 the canonical transcript was not expressed; instead three alternative transcripts were present. These transcripts differed from each other in the region encompassing exons 5 and 6. Two of these splice variants were produced by the usage of splice donor sites internal to exon 5, while a third variant was produced by retaining intron 5 in the transcript. In order to validate the presence of alternative transcripts, we performed an RT-PCR experiment with primers spanning exons 2–7 and 3–7. In Case 2 we observed three bands instead of one with all primer combinations. As control PCR we amplified the region spanning between exons 6 and 7, and we got only one band of the expected size both in the control and in Case 2 (Fig. 2C). Bands were cloned and sequenced. One fragment corresponded to an alternative transcript of SLC25A42 missing 89 bp of exon 5, the other corresponded to a variant missing 46 bp of exon 5, and the last corresponded to a band having additional 541 bp and corresponded to the variant retaining the intron 5–6 (Fig. 2D). All three alternative splice variants might potentially encode for proteins of reduced molecular weight compared to the reference protein and share with it only the N-terminus region (https://www.ncbi.nlm.nih.gov/orffinder/). The variant with the 89 bp deletion might encode a protein of 19.6 kDa and the variant with the 46 bp deletion a protein of 14.6 kDa, and the variant with the intron retained a protein of 17.6 kDa (Fig. 2D). Immunoblot analysis with three different antibodies directed versus the N-terminus, C-terminus, or whole sequence of SLC25A42 was performed in order to verify the presence of alternative protein products in the fibroblast extract of Case 2. Unfortunately, all tested antibodies failed to detect the alternative forms in Case 2 fibroblasts, as well as the canonical SLC25A42 protein not only in Case 2, as expected, but also in control fibroblasts (not shown). Probably, the low expression level of SLC25A42 in fibroblasts compared to other tissues (Supplementary Fig. 2) could explain why SLC25A42 is undetectable in total fibroblasts extracts.
Loss of SLC25A42 Is Associated with Reduced Amounts of Coenzyme A in Fibroblasts
We measured total cellular CoA in fibroblasts from Case 2 and a healthy control. We found that the total cellular CoA was reduced of a 20% in Case 2 compared to control fibroblasts. Although mild, the CoA deficiency was consistently found in all independent measurements (Fig. 3).
CoA measurement. Total cellular CoA has been measured in control and in SLC25A42-mutant fibroblasts. Values were normalized to the median of the control in each independent experiment (n = 2). p-values were calculated with an independent sample t-test. All p-values were two-sided with a significance level of 0.05
Discussion
The homozygous missense mutation c.871A>G, p.Asn291Asp in SLC25A42 was proposed as a molecular cause of mitochondrial myopathy initially in a single case of Arab descent (Shamseldin et al. 2016). Clinical features included elevated lactate levels in plasma, motor developmental delay, and proximal muscle weakness. Histological examination performed on a skeletal muscle biopsy showed ragged red-like fibers with enhanced subsarcolemmal oxidative enzyme activity as well as cytochrome c oxidase-negative fibers. Additional clinical findings included dysarthria, scoliosis, and nonprogressive myopia. Creatine kinase activity, nerve conduction velocities, brain MRI, and echocardiography were reported normal. The boy had normal cognitive function and the disease course was stable. Subsequently, 12 additional cases of Arab descent carrying the same founder mutation and presenting with a broader phenotypic spectrum characterized by the presence of developmental delay and lactic acidosis as key clinical features were reported (Almannai et al. 2018). The severity of the clinical manifestations was highly variable even within affected individuals of the same family, ranging from asymptomatic lactic acidosis to severe intellectual disability, metabolic crisis, and multiorgan involvement. In four individuals was also noticed the appearance of a movement disorder, mainly dystonia. For the first time, SLC25A42 mutations were associated with encephalopathy in several individuals.
In the current manuscript, we provide additional evidence for the implication of biallelic SLC25A42 mutations in mitochondrial diseases, as we diagnosed two additional cases with pathogenic variants in SLC25A42. Case 1, of Arab descent, carried the already described missense change c.871A>G, p.Asn291Asp, while Case 2, of European descent, carried the homozygous splice change c.380+2T>A, p.(?) as a novel disease-causing mutation. Both individuals presented with lactic acidosis, muscular hypotonia, and developmental delay as observed in the case described in Shamseldin et al. (2016) and some cases from Almannai et al. (2018), although the muscular hypotonia and cognitive impairment were more severe in Case 1 than Case 2.
Moreover, Case 1 developed also dystonia, confirming the association of SLC25A42 with movement disorders (Almannai et al. 2018). In both individuals brain MRI showed abnormalities, similarly to what observed in individuals with neurological symptoms described in Almannai et al. (2018). Our observations remark that specific mutations in SLC25A42 are likely to be identified in different ethnic populations, consistent with founder effects, and that SLC25A42 deficiency manifests not only with isolated myopathy plus lactic acidosis but includes often severe presentations with encephalomyopathy.
To further investigate the suggested role of SLC25A42 in CoA metabolism, we took advantage of available fibroblasts cell line (Case 2 only). Indeed, we observed a mild but significant reduction in the amount of total cellular CoA. We speculated that the defect might be more pronounced and functionally relevant in tissues containing more mitochondria and high SLC25A42 expression levels (brain, muscle, liver, heart, and kidney). Considering that the concentration of CoA is almost 100-fold higher in mitochondria than in the cytosol, the reduction of total cellular CoA detected in our experiments might possibly reflect the decrease in mitochondrial CoA content. How SLC25A42 impairment leads to reduced cellular CoA remains unclear, and additional experiments are required to elucidate the mechanism leading to the defect. However, the decrease in cellular CoA suggests pantethine as possible treatment for SLC25S42-affected individuals. In fact, pantethine supplementation replenished CoA levels and improved the phenotype in a Drosophila model of PKAN (PANK2-associated neurodegeneration) presenting with reduced levels of CoA and neurodegeneration (Rana et al. 2010) and rescued the viability in a Drosophila model of PPCS deficiency (Iuso et al. 2018).
In conclusion, with the identification of two additional cases carrying mutations in SLC25A42, we corroborate the association between pathogenic mutations in SLC25A42 and a heterogeneous clinical spectrum including myopathy, developmental delay, lactic acidosis, and encephalopathy. Furthermore, we highlight the functional consequences of the novel homozygous variant c.380+2T>A, suggesting a possible therapeutic approach for SLC25A42-affected individuals.
References
Almannai M, Alsamri A, Alqasmi A, Faqeih E, AlMutairi F, Alotaibi M et al (2018) Expanding the phenotype of SLC25A42-associated mitochondrial encephalomyopathy. Clin Genet 93:1097–1102
Bosveld F, Rana A, van der Wouden PE, Lemstra W, Ritsema M, Kampinga HH et al (2008) De novo CoA biosynthesis is required to maintain DNA integrity during development of the Drosophila nervous system. Hum Mol Genet 17(13):2058–2069
Dusi S, Valletta L, Haack TB, Tsuchiya Y, Venco P, Pasqualato S et al (2014) Exome sequence reveals mutations in CoA synthase as a cause of neurodegeneration with brain iron accumulation. Am J Hum Genet 94(1):11–22
Fiermonte G, Paradies E, Todisco S, Marobbio CM, Palmieri F (2009) A novel member of solute carrier family 25 (SLC25A42) is a transporter of coenzyme A and adenosine 3′,5′-diphosphate in human mitochondria. J Biol Chem 284(27):18152–18159
Haack TB, Danhauser K, Haberberger B, Hoser J, Strecker V, Boehm D et al (2010) Exome sequencing identifies ACAD9 mutations as a cause of complex I deficiency. Nat Genet 42(12):1131–1134
Haack TB, Kopajtich R, Freisinger P, Wieland T, Rorbach J, Nicholls TJ et al (2013) ELAC2 mutations cause a mitochondrial RNA processing defect associated with hypertrophic cardiomyopathy. Am J Hum Genet 93(2):211–223
Iuso A, Wiersma M, Schüller HJ, Pode-Shakked B, Marek-Yagel D, Grigat M, Schwarzmayr T, Berutti R, Alhaddad B, Kanon B, Grzechik NA, Okun JG, Perles Z, Salem Y, Barel O, Vardi A, Rubinshtein M, Tirosh T, Dubnov-Raz G, Messias AC, Terrile C, Barshack I, Volkov A, Avivi C, Eyal E, Mastantuono E, Kumbar M, Abudi S, Braunisch M, Strom TM, Meitinger T, Hoffmann GF, Prokisch H, Haack TB, Brundel BJJM, Haas D, Sibon OCM, Anikster Y (2018) Mutations in phosphopantothenoylcysteine synthetase (PPCS) cause dilated cardiomyopathy. Am J Hum Genet. https://doi.org/10.1016/j.ajhg.2018.03.022
Leonardi R, Zhang YM, Rock CO, Jackowski S (2005) Coenzyme A: back in action. Prog Lipid Res 44(2–3):125–153
Palmieri F (2004) The mitochondrial transporter family (SLC25): physiological and pathological implications. Pflugers Arch 447(5):689–709
Palmieri F (2013) The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Asp Med 34(2–3):465–484
Palmieri F (2014) Mitochondrial transporters of the SLC25 family and associated diseases: a review. J Inherit Metab Dis 37(4):565–575
Palmieri F, Monne M (2016) Discoveries, metabolic roles and diseases of mitochondrial carriers: a review. Biochim Biophys Acta 1863(10):2362–2378
Rana A, Seinen E, Siudeja K, Muntendam R, Srinivasan B, van der Want JJ et al (2010) Pantethine rescues a Drosophila model for pantothenate kinase-associated neurodegeneration. Proc Natl Acad Sci U S A 107(15):6988–6993
Shamseldin HE, Smith LL, Kentab A, Alkhalidi H, Summers B, Alsedairy H et al (2016) Mutation of the mitochondrial carrier SLC25A42 causes a novel form of mitochondrial myopathy in humans. Hum Genet 135(1):21–30
Vozza A, De Leonardis F, Paradies E, De Grassi A, Pierri CL, Parisi G et al (2017) Biochemical characterization of a new mitochondrial transporter of dephosphocoenzyme A in Drosophila melanogaster. Biochim Biophys Acta 1858(2):137–146
Acknowledgments
Study funding: TBH was supported by the German Federal Ministry of Education and Research (BMBF) within the framework of the e:Med research and funding concept (grant #FKZ 01ZX1405C). HP by the E-Rare project GENOMIT (01GM1207) and the EU Horizon 2020 Collaborative Research Project SOUND (633974). AI by the EU project TIRCON.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Additional information
Communicated by: Wolfgang Sperl, MD, PhD
Electronic Supplementary Material
Supplementary Fig. 1
Primer sequences (a), PCR conditions (b), and amplicon sizes (bp) of SLC25A42 exons 2–7, 3–7, and 6–7 (TIF 90 kb)
Supplementary Fig. 2
Expression of SLC25A42 in human tissues. Raw read counts and total amount of sequence normalized values (FPKM) indicate an about tenfold lower expression in fibroblasts compared to the brain, muscle, liver, and kidney tissues, respectively. Data were extracted from our in-house database (PDF 3 kb)
Supplementary Table 1
Clinical and genetic findings in SLC25A42-affected individuals (Modified from Almannai et al. 2018) (XLSX 14 kb)
Supplementary Table 2
Compound heterozygous and homozygous variants detected using whole exome sequencing, after applying quality and frequency filters, in Case 1 and 2, respectively (XLSX 17 kb)
Appendices
Synopsis
Mutations in SLC25A45 cause epileptic encephalopathy.
Details of the Contributions of Individual Authors
Conceived and designed the work: AI, BA, HP, and TBH. Performed the experiments: AI, CT, and EG. Analyzed and interpreted the clinical and genetic data: CW, UK, EM, TS, TMS, GFH, TM, and TBH. Drafted the article: AI, BA, and TBH. Critically revised the draft: all authors.
Compliance with Ethics Guidelines
Conflict of Interest
A. Iuso, B. Alhaddad, C. Weigel, U. Kotzaeridou, E. Mastantuono, T. Schwarzmayr, E. Graf, C. Terrile, H. Prokisch, T. M. Strom, G. F. Hoffmann, T. Meitinger, and T. B. Haack declare no conflict of interest.
Details of Ethics Approval
All procedures followed were in accordance with the ethical standards of the responsible committee on human experimentation (institutional and national) and with the Helsinki Declaration of 1975, as revised in 2000.
A Patient Consent Statement
Written informed consent was obtained from all individuals or caregivers.
Rights and permissions
Copyright information
© 2018 Society for the Study of Inborn Errors of Metabolism (SSIEM)
About this chapter
Cite this chapter
Iuso, A. et al. (2018). A Homozygous Splice Site Mutation in SLC25A42, Encoding the Mitochondrial Transporter of Coenzyme A, Causes Metabolic Crises and Epileptic Encephalopathy. In: Morava, E., Baumgartner, M., Patterson, M., Rahman, S., Zschocke, J., Peters, V. (eds) JIMD Reports, Volume 44. JIMD Reports, vol 44. Springer, Berlin, Heidelberg. https://doi.org/10.1007/8904_2018_115
Download citation
DOI: https://doi.org/10.1007/8904_2018_115
Received:
Revised:
Accepted:
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-58616-7
Online ISBN: 978-3-662-58617-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)