Abstract
The presence of posttranslational modifications is a key feature for the biological activity and stability of many natural products, including lanthipeptides. Lanthipeptides are a group of posttranslationally modified peptides containing lanthionine residues in their structures. Among lanthipeptides, the subgroup of lantibiotics is receiving a renewed interest in the last years for their high antimicrobial activity. The development of production platforms for novel lantibiotics and peptides containing posttranslational modifications is taking place in parallel to the understanding of the mechanistic aspects of the enzymes responsible for these modifications. It is now possible to combine pieces (i.e., enzymes and regulatory elements) of different biosynthesis routes of natural products in order to modify peptides of interest in a predictable manner. In this chapter we detail the production of lanthipeptides and thioether-stabilized peptides using two well-established production platforms for Escherichia coli and Lactococcus lactis. These are based on the dehydration and cyclization reactions carried out by the enzymes NisB and NisC, respectively, on peptides fused to the nisin leader peptide. Furthermore, we provide appropriate protocols for the purification and characterization of the peptides produced and modified with these biosynthetic tools.
Access provided by CONRICYT – Journals CONACYT. Download protocol PDF
Similar content being viewed by others
Keywords
1 Introduction
Lanthipeptides constitute a group of ribosomally produced and posttranslationally modified peptides (RiPPs) that display diverse activities [1, 2]. They show as a main feature the presence of lanthionine, which consists of the linkage of two amino acids (i.e., a dehydrated serine or threonine residue and a cysteine) via a thioether bond. The best characterized and studied lanthipeptides are those with antimicrobial activity, which are referred to as lantibiotics (lanthionine-containing antibiotics). Lantibiotics display activity against a wide range of Gram-positive food spoilage and pathogenic bacteria (Listeria, Streptococcus, Clostridium, Staphyloccus, and many others) [3–6]. The challenge that multidrug-resistant bacteria such as methicillin-resistant Staphylococcus aureus and vancomycin-resistant Enterococcus faecalis pose in clinics nowadays and the activity of lantibiotics against such organisms has brought them to a front position in antibiotic research [7]. Their potent antimicrobial activity, which is in the concentration range of antibiotics conventionally used in therapeutics, and the conserved activity against multidrug-resistant microorganisms clearly indicate a potential medical use [3, 8]. In fact, the scarce pharmacokinetic and pharmacodynamic studies carried out on lantibiotics point at their applicability. Furthermore, the interest of lantibiotics in therapeutics is due not only to their antimicrobial activity but also to the possibilities that their biosynthesis machinery offers for the synthesis of thioether-stabilized peptide drugs (vide infra).
The biosynthesis of lanthipeptides requires different steps. First, a serine or threonine residue is dehydrated rendering dehydroalanine (Dha) or dehydrobutyrine (Dhb), respectively. In a second step, the thiol group of a cysteine is coupled to the dehydroamino acid in a regio- and stereospecific manner to produce the lanthionine (cysteine coupled to Dha) or methyllanthionine (cysteine coupled to Dhb) ring [2]. Depending on the enzymes that catalyze these reactions, lanthipeptides are divided into four different classes. In class I two separate enzymes are responsible for the dehydration (LanB) and cyclization (LanC). In class II, III, and IV lanthipeptides, a single multidomain enzyme can do both conversions (LanM, LanKC, and LanL, respectively). The posttranslational modification (PTM) enzymes recognize specific regions within the leader peptide of the lanthipeptide and this favors the transition from an inactive state to an activated one [9–12]. Additional posttranslational modifications can take place in lanthipeptides, which increase their chemical diversity and have a great impact on their activity [13, 14].
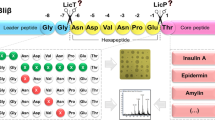
The substrate specificity of the lanthipeptide dehydratases and cyclases has been extensively studied. The modification machinery of lacticin 481 (Fig. 1) has been the most thoroughly studied LanM lantibiotic synthetase. It has been proved that the enzyme LctM can modify in vitro and in vivo diverse peptide substrates fused to the lacticin 481 leader peptide including peptides containing noncanonical amino acids [15–19]. Nevertheless, this promiscuity of the LanM synthetases does not hold true for all the studied LanMs. It has been shown that the lichenicidin and haloduracin machineries are more restricted regarding their substrate tolerance [20].
In our lab we focus on the nisin modification machinery, which belongs to the class I lantibiotics (Fig. 1). Nisin is the prototype of class I lantibiotics, and its mechanism of action, biosynthesis, regulation, and applications have been extensively studied [21–23]. The nisin gene cluster consists of 11 genes encoding the immunity proteins NisI and NisFEG; a two-component system autoinduced by nisin, NisRK; the structural peptide NisA; and the biosynthesis machinery composed of NisB (dehydratase), NisC (cyclase), NisT (transporter), and NisP (leader peptidase) [24–28]. The two-component system NisRK is widely used for the overexpression of proteins in diverse Gram-positive organisms [29, 30]. The nisin dehydratase, transporter, and cyclase have been characterized for their substrate specificity proving their broad substrate tolerance [31–33] and applications in biotechnology for extracellular protein display [34], biosynthesis of novel lantibiotics [35, 36], and the biosynthesis of lanthionine-containing peptide hormones with increased stability and potency [37, 38]. NisBTC can modify and transport different peptide sequences fused to the nisin leader peptide which functions as a recognition motif for the enzymes and at the same time keeps the modified peptide inactive until it is cleaved by proteolysis [10, 12, 27, 39, 40].
The dissection of the nisin machinery into regulatory (NisRK) and enzymatic modules (NisBTC) provides very useful tools for synthetic biology studies focused on the production of novel bioactive molecules [14]. In this way, lantibiotics present in genomes of bacteria can be mined in silico with appropriate tools such as Bagel3 [41] and expressed using the nisin biosynthesis machinery as a plug-and-play system. This basic system has been extended with other PTM enzymes from other lantibiotic gene clusters such as the reductase LtnJ from the lacticin 3147 gene cluster or the oxidative decarboxylase GdmD from the gallidermin cluster [35]. These preliminary works provide a look into the chemical diversity that can be achieved in ribosomally produced peptides using adequate enzymatic modules.
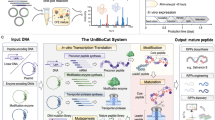
In this chapter we collect a set of techniques that allow the production and purification of novel compounds modified with the nisin modification machinery in both Escherichia coli and Lactococcus lactis. We describe a basic system for L. lactis composed of two plasmids where one is carrying the genes nisBTC under the control of the nisin-inducible promoter and a second vector contains the structural gene to be expressed (Fig. 2). This basic system has been recently extended with the insertion of a zinc-regulated promoter controlling the expression of the structural gene and the insertion of additional PTMs [35, 42]. This demonstrates the versatility of the L. lactis platform for the design and production of posttranslationally modified peptides. Similarly, we provide insight into one of the production platforms designed for E. coli, which has also dissected the PTM enzymes of the nisin cluster into two vectors with the enzymes and the structural peptide under the control of the T7 promoter (Fig. 3). We include the description of protocols for the design and construction of genes of interest encoding for the structural peptide, the induction of the expression strains, the purification of the target peptides by immobilized metal affinity chromatography or cation exchange chromatography, and the characterization of the peptide modification extent by mass spectrometry.
2 Materials
2.1 Construction of Expression Vectors for L. lactis
-
pNZE-empty [35], pNZnisA-E3 [32], pCZ-Cm [42], pIL3BTC [43], pIL3EryBTC [35] (Fig. 2).
-
Designed primers or synthetic genes encoding for the peptide of interest.
-
Phusion polymerase (Thermo Fisher Scientific, http://www.thermofisher.com) and thermal cycler.
-
Media:
-
M17 (Difco): 5.0 g pancreatic digest of casein, 5.0 g soya peptone, 5.0 g beef extract, 2.5 g yeast extract, 0.5 g ascorbic acid, 0.25 g magnesium sulfate, 19.0 g di-sodium-glycerophosphate.
-
SM17: M17 containing 0.5 M sucrose.
-
SGGM17: M17 containing 0.5 M sucrose, 0.5% glucose, 1% glycine.
-
SGM17MC: M17 containing 0.5 M sucrose, 0.5% glucose, 20 mM MgCl2, 2 mM CaCl2.
-
GSM17agar: M17 containing 0.5 M sucrose, 0.5% glucose, and 1.5% agar.
-
M17 can be purchased as a powder mixture of the different components. In our lab we use M17 from Difco. For the preparation of SM17 with or without agar, weigh the amount of powder required and add sucrose to a final concentration of 0.5 M. SM17 can be autoclaved at 121°C for 15 min and stored up to 3 months. Glucose and glycine are added from a 20% stock solution (see Note 2). CaCl2 and MgCl2 are added from 0.1 and 1 M solutions, respectively.
-
Buffers and general reagents:
-
0.5 M sucrose (see Note 3).
-
Sterile 1 M MgCl2.
-
Sterile 0.1 M CaCl2.
-
-
Antibiotics: erythromycin 5 mg/mL (1,000×) and chloramphenicol 5 mg/mL (1,000×). Dissolve the antibiotic in 70% ethanol and store at −20°C.
-
Sterile electroporation cuvettes (2 mm separation between the electrodes) and electroporator. In our lab we purchase both from Bio-Rad.
2.2 Protein Expression in L. lactis
-
Spectrophotometer.
-
Nisin stock solution – 5 μg/mL (1,000×). Mix commercial nisin powder (Sigma-Aldrich, 2.5% m/m purity) with 0.05% acetic acid (see Note 4) and shake for 1 h in a rotor. Centrifuge to remove insoluble particles and filter the supernatant. Aliquot and freeze at −20°C. Avoid freezing and thawing repeatedly, making small volume aliquots.
-
Sterile 0.5 M ZnSO4 (only required if the zinc-inducible system is used).
-
Antibiotics: erythromycin 5 mg/mL (1,000×) and chloramphenicol 5 mg/mL (1,000×). Dissolve the antibiotic in 70% ethanol and store at −20°C.
-
Media:
-
GM17: M17 containing 0.5% glucose (see Note 2).
-
Chemically defined medium (CDM). This medium is prepared according to Poolman and Konings (1988) [45] with minor modifications. It is composed of five different solutions that are mixed before use.
-
Vitamin mix (100×, composition per liter): 0.2 g nicotinic acid, 0.1 g thiamine hydrochloride, 0.1 g riboflavin, 0.1 g calcium pantothenate, 1.0 g 4-aminobenzoic acid, 1.0 g biotin, 0.1 g folic acid, 0.1 g cyanocobalamine, 0.5 g orotic acid, 0.5 g 2-deoxythymidine, 0.5 g inosine, 0.25 g DL-6,8-thioctic acid, 0.5 g pyridoxamine dihydrochloride, 0.2 g pyridoxal HCl. Mix the components, adjust the pH to 7.0, and filter-sterilize (see Note 5). Store in the dark at −20°C.
-
Amino acid mix (20×, composition per liter): 3.5 g glycine, 4.75 g l-alanine, 2.5 g l-arginine, 7.0 g l-asparagine, 7.8 g l-glutamine, 3.0 g l-histidine, 4.25 g l-isoleucine, 9.5 g l-leucine, 8.75 g l-lysine, 2.5 g l-methionine, 5.5 g l-phenylalanine, 13.5 g l-proline, 6.75 g l-serine, 4.5 g l-threonine, 1.0 g l-tryptophan, 6.5 g l-valine. Mix the amino acids, adjust the pH to 7.0, and filter-sterilize.
-
Metal mix (400×, composition per liter). Solution 1: 160.0 g MgCl2, 20.0 g CaCl2, 2.0 g ZnSO4 · 7H2O (see Note 6). Solution 2: 1.2 g CuSO4 · 5H2O, 0.08 g CoCl2. Solution 3: 2.0 g FeCl2 · 4H2O. Solution 4: 14.0 g MnSO4. Filter-sterilize each solution and keep in the dark at −20°C.
-
Base mix (100×, composition per liter): 1.0 g adenine, 1.0 g uracil, 1.0 g xanthine, 1.0 g guanine. Dissolve in milliQ water and add a few drops of 0.2 M NaOH to facilitate the solubilization. Filter-sterilize and store in the dark at −20°C.
-
Basic medium (composition per 920 mL): 0.29 g tyrosine, 13.6 g KH2PO4, 25.1 g K2HPO4, 0.6 g ammonium citrate, 1.0 g sodium acetate. Adjust the pH to 6.5, autoclave, and store in the dark at −20°C.
-
Add 10 mL of vitamin mix, 50 mL of amino acid mix, 2.5 mL of each one of the four metal solutions, and 10 mL of the base mix to 920 mL of basic medium. When this is properly mixed, add glucose at a final concentration of 0.5% and 0.25 g cysteine.
-
Minimal expression medium (MEM) [43]: similarly to CDM, MEM is prepared as separate solutions in order to facilitate the solubilization of all the components, facilitate the sterilization, and allow long-term storage. The vitamin mix is sterilized by filtration and can be frozen in aliquots at −20°C (see Note 5). The buffer and nutrient solutions are prepared separately in 80% and 20% of the final volume, respectively, and mixed before autoclaving at 121°C for 15 min. Glucose is added at a final concentration of 0.5% from a filter-sterilized 20% solution (see Note 2). The vitamins are added just before use.
-
Vitamin mix (1,000×, composition for 100 mL): 0.01 g of biotin, 0.1 g of folic acid, 0.1 g of riboflavin, 0.1 g of nicotinic acid, 0.1 g of pantothenic acid, 0.2 g of pyridoxal.
-
Buffer solution (composition per liter): 2.0 g (NH4)2SO4, 7.48 g Na2HPO4 · 2H2O, 3.0 g KH2PO4, 1.0 g NaCl.
-
Nutrient solution (composition per liter): 10.0 g casamino acids, 2.0 g sodium acetate, 0.08 g asparagine, 0.3345 g MgCl2 · 4H2O, 0.0165 g CaCl2 · 4H2O, 0.00054 g FeCl3 · 6H2O.
2.3 Construction of Lantibiotic Expression Vectors for E. coli (See Note 7)
-
Vectors for the expression of peptides containing lantibiotic modifications (see Note 8): pACYCDuet-1 and pRSFDuet-1 (EMD Millipore, http://www.emdmillipore.com).
-
PCR cleanup kit (Macherey-Nagel, http://www.mn-net.com).
-
FastDigest restriction enzymes (Thermo Fisher Scientific, http://www.thermofisher.com).
-
T4 ligase (Thermo Fisher Scientific, http://www.thermofisher.com).
-
Phusion high-fidelity DNA polymerase (Thermo Fisher Scientific, http://www.thermofisher.com).
-
Bacterial strain: E. coli DH5α (Invitrogen, http://www.lifetechnologies.com) is used for cloning. Electrocompetent cells were prepared according to standard protocols [46].
-
Antibiotics: 50 mg/mL kanamycin (dissolved in MilliQ water and filtered) and 25 mg/mL chloramphenicol (dissolved in 70% ethanol). The antibiotic stocks are aliquoted and stored at −20°C. The final concentration in the media is 50 and 25 μg/ml, respectively.
-
LB-Lennox broth (Formedium, http://www.formedium.com) (composition per liter): 10.0 g tryptone, 5.0 g yeast extract, 5.0 g NaCl. When solid media were required, 1.5% agar was added. Autoclave at 121°C for 15 min.
-
Plasmid miniprep kit (Macherey-Nagel, http://www.mn-net.com).
2.4 Expression of Peptides Containing Lantibiotic Modifications in E. coli
-
Expression vectors: pRSFDuet-1 containing nisA and nisB and pACYCDuet-1 containing nisC (Fig. 3).
-
Terrific broth (Formedium, http://www.formedium.com) (composition per liter): 12.0 g tryptone, 24.0 g yeast extract, 4.0 ml glycerol (see Note 9). Autoclave at 121°C for 15 min.
-
Antibiotics: 50 mg/mL kanamycin and 25 mg/mL chloramphenicol. Dissolve kanamycin in water (filtering is required) and chloramphenicol in 70% ethanol. Aliquot and store at −20°C. The final concentration in the medium is 50 μg/mL for kanamycin and 25 μg/mL for chloramphenicol.
-
Expression strain: E. coli BL21(DE3) (Invitrogen, http://www.lifetechnologies.com) (see Note 10).
-
Isopropyl β-d-1-thiogalactopyranoside (IPTG): 1 M in milliQ water. Filter-sterilize and store at −20°C.
-
Sonication lysis buffer (http://www.embl.de): 50 mM TrisHCl, pH 7.5, 200 mM NaCl, 5 mM DTT, 1 mM phenylmethylsulfonyl fluoride (PMSF).
-
Ultrasonic homogenizer (Sonics Vibra-Cell VCX130, www.sonics.com). Optionally a French press could be used to disrupt cells.
2.5 Purification of Lanthionine-Containing Peptides Using Cationic Exchange Chromatography
-
Dilution buffer: 100 mM lactate.
-
Wash buffer: 50 mM lactate pH 4.0 (see Note 11).
-
Elution buffer: 50 mM lactate 1 M NaCl pH 4.0.
-
Storage buffer: 0.2 M sodium acetate + 20% ethanol.
-
75% acetic acid.
-
2 M NaCl.
-
1 M NaOH.
-
HiTrap SP HP 5 mL prepacked column (GE Healthcare, http://www.gelifesciences.com).
Prepare all the solutions according to the recipe using MilliQ water and filter them with a 0.45 μm filter to remove any particles that can damage the column.
2.6 Purification of Lantibiotics on Nickel-NTA Column
-
Ni-NTA resin (Qiagen, http://www.qiagen.com).
-
Lysis buffer: 50 mM NaH2PO4, 300 mM NaCl, 10 mM imidazole, pH 7.0.
-
Wash buffer: 50 mM NaH2PO4, 300 mM NaCl, 20 mM imidazole, pH 7.0.
-
Elution buffer: 50 mM NaH2PO4, 300 mM NaCl, 250 mM imidazole, pH 7.0.
-
20% ethanol.
2.7 Salt Removal of Semipurified Lanthionine-Containing Peptide
-
MilliQ water.
-
PD-10 G-25 Sephadex prepacked gel filtration columns (GE Healthcare, http://www.gelifesciences.com).
2.8 Cleavage of the Leader Peptide
-
Sequencing-grade trypsin (Sigma-Aldrich, www.sigma-aldrich.com) dissolved in onefold cleavage buffer.
-
Tenfold cleavage buffer: 50 mM CaCl2 500 mM Tris pH 6.5.
2.9 Reversed-Phase HPLC Separation of the Lanthionine-Containing Peptide
-
Chromatographer: we use a semipreparative Agilent 1200 series HPLC (www.agilent.com).
-
Solvent A: 0.1% trifluoroacetic acid in MilliQ water.
-
Solvent B: acetonitrile 0.1% trifluoroacetic acid.
-
Analytical C-12 column Jupiter Proteo 90Å, 4 μm, 250 × 4.6 mm, with a precolumn of the same material (Phenomenex, www.phenomenex.com).
-
Semipreparative C-12 column Jupiter Proteo 90Å, 4 μm, 250 × 10 mm, with a precolumn of the same material (Phenomenex, www.phenomenex.com).
2.10 Determination of the Peptide Mass Using MALDI MS
-
Mass spectrometer: Applied Biosystems Voyager DE Pro mass spectrometer.
-
50% acetonitrile, 0.1% trifluoroacetic acid.
-
Matrix: α-cyano-4-hydroxy-cinnamic acid. Dissolve the matrix in 50% acetonitrile 0.1% trifluoroacetic acid at a final concentration of 5 mg/mL (see Note 12).
-
Peptide standard (Sigma-Aldrich): ACTH fragment (2,465.19 Da), insulin oxidized β-chain (3,494.65 Da), bovine insulin (5,735.00 Da). Dissolve each peptide standard in 100 μL of 50% acetonitrile 0.1% trifluoroacetic acid to obtain a 100 pmol/μL solution and mix the three solutions (final concentration 33.3 pmol/μl of each peptide). Aliquot in small volumes (we use 10 μL per tube) to prevent repeated thawing and freezing and store at −20°C.
3 Methods
3.1 Protocol for the Construction of Lantibiotic Expression Vectors in L. lactis
3.1.1 Preparation of Competent Cells [47]
-
1.
Inoculate L. lactis NZ9000 in SGGM17 (see Note 13) and incubate overnight at 30°C.
-
2.
Use the overnight culture to inoculate fresh SGGM17 at a final concentration of 4% (see Note 14).
-
3.
Grow the cells until they reach an OD600nm of 0.4–0.6.
-
4.
Centrifuge the culture at 8,000 × g for 10 min in a precooled centrifuge at 4°C (see Note 15).
-
5.
Discard the supernatant and carefully resuspend the cells in 0.5 volumes of ice-cold 0.5 M sucrose containing 10% glycerol. Avoid vortexing. Centrifuge in the same conditions as step 4.
-
6.
Repeat step 5 two more times.
-
7.
Resuspend the cell pellet in 0.01 volumes of ice-cold 0.5 M sucrose containing 10% glycerol.
-
8.
Prepare 40 μL aliquots in sterile cold tubes and freeze them at −80°C if they are not going to be used immediately (see Note 16).
3.1.2 Transformation
-
1.
Mix the plasmid DNA with an aliquot of competent cells pipetting smoothly (see Note 17). Leave on ice for 1 min.
-
2.
Transfer the cells to an ice-cold electroporation cuvette (see Note 18).
-
3.
Place the cuvette in the electroporator and apply an electrical pulse at 200 Ω, 2.5 kV, 25 μF.
-
4.
Immediately after the pulse add 1 mL of SGM17MC.
-
5.
Keep on ice for 5 min.
-
6.
Incubate 2 h at 30°C.
-
7.
Plate the cells on SGM17agar with the appropriate antibiotic.
3.1.3 Construction of a Peptide Fused to Nisin Leader Peptide
The expression of lantibiotics, dehydrated and thioether-stabilized peptides in L. lactis, is based on a two plasmids system (Fig. 2). The first one is a pIL253-based vector that can harbor large inserts and carries the PTM enzymes NisB, NisC, and NisT. The second plasmid is a pSH71-based vector that can replicate in diverse bacteria including E. coli and L. lactis. Although the pSH71-derived vector can serve as a shuttle vector, the easy manipulation and transformation of L. lactis using electroporation in an osmotically controlled environment make the intermediate cloning in E. coli not necessary and saves time (see previous protocol).
In either case, the control of the expression is achieved with the use of the nisin-controlled expression system (NICE). This system requires the insertion of a nisin-inducible promoter (either P nisA or P nisF ) in front of the gene(s) of interest and the presence of NisRK in the producer strain to sense the addition of nisin. NisRK can be encoded in an additional plasmid or in the chromosome. Due to the presence of two plasmids in the system, we recommend the use of strains with nisRK integrated in the chromosome such as L. lactis NZ9000, which is an MG1363 derivative with nisRK integrated in the pepN locus [48]. A recently developed system includes in addition to the nisin-inducible system a zinc-inducible promoter so that the enzymes and the structural peptide can be induced separately [42]. This is an advantage when additional posttranslational modification enzymes are included in the system and induction timing needs to be controlled.
3.1.3.1 Cloning a Synthetic Construct
The existing vectors containing the gene nisA or its derivatives do not contain restriction sites in their sequence that allow the cloning of small fragments fused to the leader peptide. Considering the small size of the genes encoding the core peptide to be fused to the nisin leader peptide, we suggest ordering synthetic constructs encompassing the promoter, the leader peptide, and the core peptide. This facilitates the cloning process and selection.
-
1.
Design the synthetic DNA containing a BglII site, the promoter P nisA , and the ORF encoding the nisin leader peptide and the core peptide, followed by a HindIII site.
-
2.
Digest the synthetic DNA and the vector pNZE-empty with BglII and HindIII.
-
3.
Ligate the vector and the DNA.
-
4.
Transform the ligation mixture into L. lactis NZ9000.
-
5.
Select positive clones by colony PCR with the primers pNZEmf (CAATTCCTTAAAACATGCAGG) and pNZrev (CAATCAAAGCAACACGTGC).
-
6.
Transform a right clone into L. lactis NZ9000 (pIL3BTC) and select transformants on SGM17-agar containing 5 μg/ml erythromycin and 5 μg/ml chloramphenicol.
3.1.3.2 Cloning a Small Peptide Sequence
-
1.
Design a reverse primer that hybridizes with the nisin leader peptide coding sequence and carries a tail coding for the peptide of interest and a phosphorylated forward primer that hybridizes with the region downstream the nisA gene in pNZnisA-E3 (Fig. 2).
-
2.
Perform a round PCR that amplifies the whole vector pNZnisA-E3 using a high-fidelity polymerase such as Phusion (Thermo Fisher Scientific. http://www.thermofisher.com).
-
3.
Clean the PCR product with an appropriate kit. We use the Macherey-Nagel kit (Macherey-Nagel, http://www.mn-net.com).
-
4.
Ligate the PCR product to circularize the amplified vector.
-
5.
Transform the ligation mixture into L. lactis NZ9000 and select transformants on SGM17-agar containing 5 μg/ml erythromycin.
-
6.
Select positive clones by colony PCR with the primers pNZEmf (CAATTCCTTAAAACATGCAGG) and pNZrev (CAATCAAAGCAACACGTGC).
-
7.
Transform a right clone into L. lactis NZ9000 (pIL3BTC) and select transformants on SGM17-agar containing 5 μg/ml erythromycin and 5 μg/ml chloramphenicol.
3.2 Protocol for Protein Induction in L. lactis
-
1.
Inoculate L. lactis NZ9000 containing the pIL3BTC and the pNZE-derivative where the gene of interest is cloned in GM17 containing 5 μg/ml erythromycin and 5 μg/ml chloramphenicol. Grow overnight at 30°C (see Note 19).
-
2.
Mix the components of the expression medium (MEM for nisin-induced expression or CDM if zinc induction is used) and prewarm them at 30°C.
-
3.
Inoculate the expression medium with the overnight culture at a final concentration of 4%.
-
4.
Grow the cells until an OD600nm between 0.4 and 0.6.
-
5.
Induce the culture with either 5 ng/mL nisin or 5 ng/mL nisin and 1 mM ZnSO4.
-
6.
Incubate 3 h at 30°C.
-
7.
Centrifuge the culture at 8,000 × g for 10 min at 4°C.
-
8.
Retain the supernatant.
3.3 Construction of Lantibiotic Expression Vectors in E. coli (See Note 7)
The expression vector pRSFDuet-1 contains a 6xHis tag in the beginning of the first polylinker between NcoI and BamHI restriction sites. This enables the introduction of 6xHis tag before the nisin leader peptide and the peptide to be expressed. Here we present the expression system with the nisin biosynthesis genes; however it is not the only possible version (see Note 8).
All PCR and DNA restriction products have to be cleaned with the PCR cleanup kit to ensure efficient further manipulations (e.g., ligation or restriction).
-
1.
Amplify the genes of choice (nisA, nisB, and nisC). When employing PCR, use high-fidelity polymerase, e.g., Phusion (see Subheading 2.3), and follow the manufacturer’s instructions.
-
2.
Digest nisA DNA fragment with BamHI and HindIII and ligate it into previously BamHI/HindIII digested pRSFDuet-1. For the ligation use T4 ligase and follow manufacturer’s instructions.
-
3.
Transform the ligation mix into E. coli DH5α and plate on LB-kanamycin agar plates (see Subheading 2.4). Incubate plates at 37°C for 16 h or until visible colonies appear.
-
4.
Check approximately 10 colonies for positive clones by colony PCR. Choose a positive clone and inoculate it into LB-kanamycin medium (see Subheading 2.4). Incubate overnight at 37°C with vigorous shaking.
-
5.
Pellet the overnight culture and use the plasmid miniprep kit to isolate the plasmid. Verify if cloning was successful by sequencing.
-
6.
Clone nisB DNA fragment into the obtained pRSFDuet-1 nisA containing vector using NdeI and XhoI restriction enzymes. Follow the instructions from steps 2–5.
-
7.
Clone nisC DNA fragment into pACYCDuet-1 vector using BglII and XhoI restriction enzymes and following the instructions from the previous steps 2–5. LB-chloramphenicol agar plates (see Subheading 2.4) must be used instead LB-kanamycin ones for selection.
3.4 Expression of Lanthionine-Containing Peptides in E. coli
The methods presented here are based on previous methodology but with minor modifications [49].
-
1.
Transform E. coli BL21(DE3) with both pRSFDuet-1 containing nisA and nisB and pACYCDuet-1 containing nisC and plate on LB-agar plates containing kanamycin and chloramphenicol (see Note 20). Incubate plates at 37°C for 16 h or until visible colonies appear.
-
2.
Grow an overnight culture from a single colony.
-
3.
Inoculate terrific broth (see Note 9) containing kanamycin and chloramphenicol (see Subheading 2.4). Grow cells at 37°C with vigorous shaking until OD600nm reaches 0.4–0.6.
-
4.
Induce the culture with 1 mM IPTG and continue growing cells at 18°C for an additional 16 h (see Note 21).
-
5.
Pellet the cells by centrifugation at 4°C, 8,000 × g for 15 min. Resuspend the cell pellet in 0.2 culture volumes of sonication buffer (see Subheading 2.4). It is possible to store the cell suspension at −20°C for several weeks; otherwise proceed with cell lysis as described in the next steps.
-
6.
Chill the suspension on ice. Lyse the cells using an ultrasonic homogenizer. Use 10 times 10 s pulses with a pulse intensity of 65% leaving 30 s intervals in between for cooling.
-
7.
Centrifuge the lysate at 18,000 × g for 30 min at 4°C.
-
8.
Collect the supernatant and discard the cell pellet.
3.5 Protocol for Cation Exchange Chromatography (See Note 22)
-
1.
Dilute the supernatant with 1 volume of dilution buffer and filter through a 0.45 μm membrane.
-
2.
Wash the HiTrap SP HP column with 5 column volumes of wash buffer.
-
3.
Wash the column with 5 column volumes of elution buffer.
-
4.
Wash the column with 5 column volumes of wash buffer.
-
5.
Run the supernatant through the column (see Note 23).
-
6.
Wash the column with 5 column volumes of wash buffer.
-
7.
Elute the peptide with elution buffer. The first 4 mL fraction corresponding to the column dead volume can be discarded. Collect the next 8 mL.
-
8.
Regenerate the column:
-
(a)
Wash with 3 volumes of MilliQ water.
-
(b)
Wash with 3 volumes of 1 M NaOH.
-
(c)
Wash with 3 volumes of MilliQ water.
-
(d)
Wash with 3 volumes of 2 M NaCl.
-
(e)
Wash with 3 volumes of MilliQ water.
-
(f)
Wash with 3 volumes of 75% acetic acid.
-
(g)
Wash with 3 volumes of MilliQ water.
-
(h)
Wash with 5 volumes of storage buffer.
-
(a)
Keep a sample of steps 1, 5, and 7 to monitor the production and purification process either by Tricine-SDS-PAGE [50] or antimicrobial tests [35] if the lanthionine-containing peptide has inhibitory activity.
3.6 Purification of Lantibiotics by Immobilized Metal Affinity Chromatography on a Nickel-NTA Column
The method presented here is based on [39].
-
1.
Equilibrate 1 mL of 50% superflow Ni-NTA resin (see Note 24) with 10 volumes of lysis buffer (see Subheading 2.6) in a 50 mL tube by mixing on a rotor for 30 min.
-
2.
Let the resin settle down, remove the buffer, and repeat step 1.
-
3.
Resuspend the column material in 4–8 mL cytoplasmic fraction obtained previously (protocol 3.4) and transfer it into a 15 mL tube. Add lysis buffer to a final volume of 12 mL.
-
4.
Allow the His-tagged peptide to bind to the column material on a rotor at 4°C for 2 h. After binding transfer the column material to a gravity column.
-
5.
Wash the column twice with 10 volumes of wash buffer (see Subheading 2.6).
-
6.
Elute the peptide with 10 column volumes of elution buffer taking fractions of approximately 0.5 mL (see Subheading 2.6).
-
7.
Wash the column with 10 volumes 20% ethanol for storage.
Take a sample of the lysate before applying the Ni-NTA resin and from steps 4 and 5 to monitor the production and purification process by Tricine-SDS-PAGE [50] or antimicrobial tests [35] if the lanthionine-containing peptide has inhibitory activity.
3.7 Desalting Protocol
-
1.
Wash the PD-10 column with 25 mL MilliQ water.
-
2.
Apply up to 2.5 mL sample and discard the flow-through.
-
3.
Elute the peptide with 3.5 mL MilliQ water.
-
4.
Repeat steps 1–3 to desalt all the peptide fractions obtained from either cation exchange chromatography or immobilized metal affinity chromatography.
3.8 Cleavage of the Leader Peptide
-
1.
Mix 9 volumes of peptide solution with 1 volume of tenfold cleavage buffer.
-
2.
Add 1 μg of sequencing-grade trypsin (see Note 25).
-
3.
Incubate at 37°C for 2 h (see Note 25).
3.9 HPLC Separation of the Lanthionine-Containing Peptide
-
1.
Equilibrate the column with 10 column volumes of 20% solvent B (see Subheading 2.9).
-
2.
Inject the digestion mix.
-
3.
Run the separation gradient:
-
(a)
Analytical column: 8 min 20% solvent B, 13 min 25% solvent B, 38 min 60% solvent B, 39 min 95% solvent B, 44 min 95% solvent B, 45 min 20% solvent B. Use a constant flow at 1 mL/min.
-
(b)
Semipreparative column: 8 min 20% solvent B, 13 min 25% solvent B, 38 min 60% solvent B, 39 min 95% solvent B, 44 min 95 % solvent B, 45 min 20 % solvent B. Use a constant flow at 2.5 mL/min.
-
(a)
-
4.
Collect the peaks for further analysis or storage.
-
5.
Remove the solvent from the sample (e.g., freeze-dryer) and store at −20°C (see Note 26).
3.10 Protocol for Mass-Spectrometry Determination Using MALDI-TOF
-
1.
Dissolve the matrix.
-
2.
Wash the MALDI target with 50% acetonitrile 0.1% trifluoroacetic acid and dry.
-
3.
Spot 1 μl of sample or protein standard mix on the target and let it dry.
-
4.
Optional: if the samples contain salts (this is not necessary for HPLC purified samples or protein standard), it is possible to use zip-tip C18 (Millipore) before spotting them on the target or wash with MilliQ water after spotting. To wash with MilliQ water, spot 5 μL MilliQ water on the sample once that it is dried and immediately remove it with a tissue by capillarity. The results are usually better with zip-tip treatment.
-
5.
Spot 1 μL of the dissolved matrix on the sample and let it dry.
-
6.
Determine the mass of the peptides in the mass spectrometer. We use 20,000 V acceleration, 94% grid, 0.05% guide wire, 100 ns delay time in linear mode acquisition.
-
7.
Calibrate the mass data with appropriate software using the peptide standard as a reference. We use external calibration and Data Explorer 4.0.0.0 (Applied Biosystems) for the analysis.
-
8.
Compare the mass values determined experimentally with the theoretical values (see Note 27).
4 Notes
-
1.
The induction in L. lactis using nisin requires the two-component system NisRK, which is integrated in the genome of strain NZ9000.
-
2.
Glucose cannot be autoclaved together with M17 or SM17; therefore filter sterilization of a 20% concentrated stock is recommended.
-
3.
It is advisable to use a fresh solution and not store it for longer than 1 month. Use MilliQ water for its preparation to minimize the presence of ions that can prevent the transformation.
-
4.
Nisin is more soluble and stable at acidic pH. It is less stable at neutral pH and unstable at alkaline pH. The commercial powder contains a high proportion of insoluble particles that have to be removed.
-
5.
The vitamin mix is sensitive to light. Store protected from light.
-
6.
Prepare the metal mix without zinc if the zinc-inducible system for L. lactis is going to be used.
-
7.
The lantibiotic expression system in E. coli briefly described here is based on Shi and coworkers methodology with some modifications [49].
-
8.
Other optional vectors could be used for heterologous lantibiotic production in E. coli: (a) single plasmid pET28a, pET28b, or pRSFDuet-1 [51]; (b) single fosmid (e.g., pCC2FOS) or in combination with other expression vectors (e.g., pET24a(+) [52]); (c) pRSFDuet-1 in combination with pCDFDuet-1 [53].
-
9.
LB could be considered to be used instead of terrific broth; however it might affect the protein production levels.
-
10.
Different expression strains might give different lantibiotic production levels (e.g., lower nisin amounts are produced in E. coli Rosetta-gami 2(DE3)) (EMD Millipore, http://www.emdmillipore.com) compared to E. coli BL21(DE3) when using the same experimental setup (unpublished results Buivydas A, Moll GN, Kuipers OP).
-
11.
In order to increase the storage time of this solution, we recommend the preparation of 0.5 M lactic acid pH 4.0 and dilute it tenfold just before use.
-
12.
α-Cyano-4-hydroxy-cinnamic acid is light- and oxygen-sensitive. Therefore, prepare it fresh, and after mixing the power with the solvent, cover with aluminum foil.
-
13.
Antibiotics can be added when competent cells containing already one plasmid are being used.
-
14.
Due to the presence of glycine in the medium, the growth speed is reduced. Thus, we recommend using prewarmed medium at 30°C to avoid additional delay.
-
15.
It is important that the cells are in mid-exponential phase to obtain optimal competence. Therefore, it is necessary chilling them and work all the time on ice and with ice-cold media and material (e.g., plastic tubes, cuvettes, etc.) from this point.
-
16.
The transformation efficiency can be improved if the cells are frozen in liquid nitrogen. However, it is not essential. Normal efficiency values of 104–105 CFU/μg DNA can be achieved using this procedure.
-
17.
If the cells were frozen, they must be thawed on ice. Do not add the DNA on frozen cells and keep the DNA volume below 4 μL. The DNA sample must contain a low salt concentration. The usual elution buffers in commercial DNA extraction kits have low salt concentration and can be used directly. Otherwise desalt your sample by precipitation or dialysis against MilliQ water on a 0.22 μm filter membrane (Millipore) for 20–60 min.
-
18.
The separation gap between the electrodes for transformation of L. lactis is 2 mm. Make sure there are no air bubbles in the cells after placing them in the cuvette and dry the outside electrode with a tissue.
-
19.
In the system using nisin and zinc as inducers, the selected vectors are pIL3EryBTC and a pCZ-Cm derivative containing the structural genes [35, 42].
-
20.
We recommend transforming E. coli BL21(DE3) simultaneously with the two plasmids by electroporation, instead of preparing competent cells containing already one of the plasmids.
-
21.
Induced cultures could be incubated, alternatively, at 37°C. However the protein production level might be lower, though sufficient for antimicrobial activity tests, MS, and/or Western blot analyses.
-
22.
This protocol is designed for lantibiotics, which have a pI above 6.0. If the pI of the produced protein is lower, anionic exchange chromatography is advised instead of reducing the pH of the solutions in this protocol.
-
23.
We use a peristaltic pump to load the column at a flow rate of 5 mL/min.
-
24.
The volume of 50% superflow Ni-NTA column resin is used in ratio 1:4 with cleared cell lysate (see Subheading 3.4). 4 ml of a lysate could be obtained from 20 to 200 mL of initial cell culture volume depending on the expression level of 6xHis-tagged protein.
-
25.
Trypsin cleaves preferentially in the arginine residue located at the end of the nisin leader peptide. Lanthionine rings confer protection of the peptide against proteolysis, reducing the possibilities of degradation. However, it might be necessary to reduce the incubation time and enzyme to peptide ratio if positively charged residues are exposed to cleavage in a particular peptide of interest.
-
26.
It is preferable storing a dried sample rather than keeping it in organic solvent. In the latter case, store at −80°C and not −20°C.
-
27.
During data analysis it is crucial to calculate the mass differences introduced by the posttranslational modification(s) taking place. For instance, each dehydration in the peptide causes a reduction of 18 Da in the mass, whereas reduction of dehydroalanine by a LanJ enzyme increases the mass 2 Da. There are no devoted tools developed for this purpose; thus the data analysis needs to be precise.
References
Arnison PG, Bibb MJ, Bierbaum G et al (2013) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160
Knerr PJ, van der Donk WA (2012) Discovery, biosynthesis, and engineering of lantipeptides. Annu Rev Biochem 81:479–505
Dischinger J, Basi Chipalu S, Bierbaum G (2014) Lantibiotics: promising candidates for future applications in health care. Int J Med Microbiol 304:51–62
Mota-Meira M, LaPointe G, Lacroix C, Lavoie MC (2000) MICs of mutacin B-Ny266, nisin A, vancomycin, and oxacillin against bacterial pathogens. Antimicrob Agents Chemother 44:24–29
Cotter PD, Ross RP, Hill C (2013) Bacteriocins – a viable alternative to antibiotics? Nat Rev Microbiol 11:95–105
Cotter PD, Hill C, Ross RP (2005) Bacteriocins: developing innate immunity for food. Nat Rev Microbiol 3:777–788
Boucher HW, Talbot GH, Bradley JS et al (2009) Bad bugs, no drugs: no ESKAPE! An update from the Infectious Diseases Society of America. Clin Infect Dis 48:1–12
Van Heel AJ, Montalban-Lopez M, Kuipers OP (2011) Evaluating the feasibility of lantibiotics as an alternative therapy against bacterial infections in humans. Expert Opin Drug Metab Toxicol 7:675–680
Oman TJ, van der Donk WA (2010) Follow the leader: the use of leader peptides to guide natural product biosynthesis. Nat Chem Biol 6:9–18
Abts A, Montalban-Lopez M, Kuipers OP, Smits SH, Schmitt L (2013) NisC binds the FxLx motif of the nisin leader peptide. Biochemistry 52:5387–5395
Plat A, Kuipers A, Rink R, Moll GN (2013) Mechanistic aspects of lanthipeptide leaders. Curr Protein Pept Sci 14(2):85–96
Plat A, Kluskens LD, Kuipers A, Rink R, Moll GN (2011) Requirements of the engineered leader peptide of nisin for inducing modification, export, and cleavage. Appl Environ Microbiol 77:604–611
Willey JM, van der Donk WA (2007) Lantibiotics: peptides of diverse structure and function. Annu Rev Microbiol 61:477–501
Montalbán-López M, Zhou L, Buivydas A, van Heel AJ, Kuipers OP (2012) Increasing the success rate of lantibiotic drug discovery by Synthetic Biology. Expert Opin Drug Discovery 7:695–709
Chatterjee C, Patton GC, Cooper L, Paul M, van der Donk WA (2006) Engineering dehydro amino acids and thioethers into peptides using lacticin 481 synthetase. Chem Biol 13:1109–1117
Furgerson Ihnken LA, Chatterjee C, van der Donk WA (2008) In vitro reconstitution and substrate specificity of a lantibiotic protease. Biochemistry 47:7352–7363
Levengood MR, van der Donk WA (2008) Use of lantibiotic synthetases for the preparation of bioactive constrained peptides. Bioorg Med Chem Lett 18:3025–3028
Levengood MR, Knerr PJ, Oman TJ, van der Donk WA (2009) In vitro mutasynthesis of lantibiotic analogues containing nonproteinogenic amino acids. J Am Chem Soc 131:12024–12025
Zhang X, van der Donk WA (2007) On the substrate specificity of dehydration by lacticin 481 synthetase. J Am Chem Soc 129:2212–2213
Caetano T, Barbosa J, Möesker E, Süssmuth RD, Mendo S (2014) Bioengineering of lanthipeptides in Escherichia coli: assessing the specificity of lichenicidin and haloduracin biosynthetic machinery. Res Microbiol 165:600–604
Breukink E, Wiedemann I, van Kraaij C, Kuipers OP, Sahl H, de Kruijff B (1999) Use of the cell wall precursor lipid II by a pore-forming peptide antibiotic. Science 286:2361–2364
Hasper HE, Kramer NE, Smith JL et al (2006) An alternative bactericidal mechanism of action for lantibiotic peptides that target lipid II. Science 313:1636–1637
Lubelski J, Rink R, Khusainov R, Moll GN, Kuipers OP (2008) Biosynthesis, immunity, regulation, mode of action and engineering of the model lantibiotic nisin. Cell Mol Life Sci 65:455–476
Siegers K, Entian KD (1995) Genes involved in immunity to the lantibiotic nisin produced by Lactococcus lactis 6F3. Appl Environ Microbiol 61:1082–1089
Kuipers OP, Beerthuyzen MM, Siezen RJ, De Vos WM (1993) Characterization of the nisin gene cluster nisABTCIPR of Lactococcus lactis. Requirement of expression of the nisA and nisI genes for development of immunity. Eur J Biochem FEBS 216:281–291
Kuipers OP, Beerthuyzen MM, de Ruyter PG, Luesink EJ, de Vos WM (1995) Autoregulation of nisin biosynthesis in Lactococcus lactis by signal transduction. J Biol Chem 270:27299–27304
Van der Meer JR, Polman J, Beerthuyzen MM, Siezen RJ, Kuipers OP, De Vos WM (1993) Characterization of the Lactococcus lactis nisin A operon genes nisP, encoding a subtilisin-like serine protease involved in precursor processing, and nisR, encoding a regulatory protein involved in nisin biosynthesis. J Bacteriol 175:2578–2588
Siezen RJ, Kuipers OP, de Vos WM (1996) Comparison of lantibiotic gene clusters and encoded proteins. Antonie Van Leeuwenhoek 69:171–184
De Ruyter PG, Kuipers OP, Beerthuyzen MM, van Alen-Boerrigter I, de Vos WM (1996) Functional analysis of promoters in the nisin gene cluster of Lactococcus lactis. J Bacteriol 178:3434–3439
Eichenbaum Z, Federle MJ, Marra D et al (1998) Use of the lactococcal nisA promoter to regulate gene expression in Gram-positive bacteria: comparison of induction level and promoter strength. Appl Environ Microbiol 64:2763–2769
Kluskens LD, Kuipers A, Rink R et al (2005) Post-translational modification of therapeutic peptides by NisB, the dehydratase of the lantibiotic nisin. Biochemistry 44:12827–12834
Kuipers A, de Boef E, Rink R et al (2004) NisT, the transporter of the lantibiotic nisin, can transport fully modified, dehydrated, and unmodified prenisin and fusions of the leader peptide with non-lantibiotic peptides. J Biol Chem 279:22176–22182
Rink R, Kluskens LD, Kuipers A, Driessen AJM, Kuipers OP, Moll GN (2007) NisC, the cyclase of the lantibiotic nisin, can catalyze cyclization of designed nonlantibiotic peptides. Biochemistry 46:13179–13189
Bosma T, Kuipers A, Bulten E, de Vries L, Rink R, Moll GN (2011) Bacterial display and screening of posttranslationally thioether-stabilized peptides. Appl Environ Microbiol 77:6794–6801
Van Heel AJ, Mu D, Montalbán-López M, Hendriks D, Kuipers OP (2013) Designing and producing modified, new-to-nature peptides with antimicrobial activity by use of a combination of various lantibiotic modification enzymes. ACS Synth Biol 2:397–404
Majchrzykiewicz JA, Lubelski J, Moll GN et al (2010) Production of a class II two-component lantibiotic of Streptococcus pneumoniae using the class I nisin synthetic machinery and leader sequence. Antimicrob Agents Chemother 54:1498–1505
Kluskens LD, Nelemans SA, Rink R et al (2009) Angiotensin-(1-7) with thioether bridge: an angiotensin-converting enzyme-resistant, potent angiotensin-(1-7) analog. J Pharmacol Exp Ther 328:849–854
Rink R, Arkema-Meter A, Baudoin I et al (2010) To protect peptide pharmaceuticals against peptidases. J Pharmacol Toxicol Methods 61:210–218
Khusainov R, Heils R, Lubelski J, Moll GN, Kuipers OP (2011) Determining sites of interaction between prenisin and its modification enzymes NisB and NisC. Mol Microbiol 82:706–718
Khusainov R, Moll GN, Kuipers OP (2013) Identification of distinct nisin leader peptide regions that determine interactions with the modification enzymes NisB and NisC. FEBS Open Bio 3:237–242
Van Heel AJ, de Jong A, Montalbán-López M, Kok J, Kuipers OP (2013) BAGEL3: automated identification of genes encoding bacteriocins and (non-)bactericidal posttranslationally modified peptides. Nucleic Acids Res 41:W448–W453
Mu D, Montalbán-López M, Masuda Y, Kuipers OP (2013) Zirex: a novel zinc-regulated expression system for Lactococcus lactis. Appl Environ Microbiol 79:4503–4508
Rink R, Kuipers A, de Boef E et al (2005) Lantibiotic structures as guidelines for the design of peptides that can be modified by lantibiotic enzymes. Biochemistry 44:8873–8882
Kuipers OP, de Ruyter PG, Kleerebezem M, de Vos WM (1997) Controlled overproduction of proteins by lactic acid bacteria. Trends Biotechnol 15:135–140
Poolman B, Konings WN (1988) Relation of growth of Streptococcus lactis and Streptococcus cremoris to amino acid transport. J Bacteriol 170:700–707
Sambrook J, Russell D, Sambrook J (2001) Molecular cloning: a laboratory manual. Cold Spring Harbor Laboratory Press, New York
Holo H, Nes IF (1989) High-frequency transformation, by electroporation, of Lactococcus lactis subsp. cremoris grown with glycine in osmotically stabilized media. Appl Environ Microbiol 55:3119–3123
De Ruyter PG, Kuipers OP, De Vos WM (1996) Controlled gene expression systems for Lactococcus lactis with the food-grade inducer nisin. Appl Environ Microbiol 62:3662–3667
Shi Y, Yang X, Garg N, van der Donk WA (2011) Production of lantipeptides in Escherichia coli. J Am Chem Soc 133:2338–2341
Schägger H (2006) Tricine-SDS-PAGE. Nat Protoc 1:16–22
Lin Y, Teng K, Huan L, Zhong J (2011) Dissection of the bridging pattern of bovicin HJ50, a lantibiotic containing a characteristic disulfide bridge. Microbiol Res 166:146–154
Caetano T, Krawczyk JM, Mösker E, Süssmuth RD, Mendo S (2011) Heterologous expression, biosynthesis, and mutagenesis of type II lantibiotics from Bacillus licheniformis in Escherichia coli. Chem Biol 18:90–100
Shi Y, Bueno A, van der Donk WA (2012) Heterologous production of the lantibiotic Ala(0)actagardine in Escherichia coli. Chem Commun (Camb) 48:10966–10968
Acknowledgments
Andrius Buivydas and Manuel Montalban-Lopez are supported by the Nederlandse Organisatie voor Wetenschappelijk Onderzoek (NWO) projects ALW 821.02.018 and 855.01.162, respectively. M. Montalban-Lopez is also part of the EU Project Synpeptide.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2015 Springer-Verlag Berlin Heidelberg
About this protocol
Cite this protocol
Montalban-Lopez, M., Buivydas, A., Kuipers, O.P. (2015). Purification of Peptide Antimicrobials and Thioether-Stabilized Molecules Produced In Vivo by Lantibiotic Modification Machineries. In: McGenity, T., Timmis, K., Nogales Fernández, B. (eds) Hydrocarbon and Lipid Microbiology Protocols. Springer Protocols Handbooks. Springer, Berlin, Heidelberg. https://doi.org/10.1007/8623_2015_122
Download citation
DOI: https://doi.org/10.1007/8623_2015_122
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-662-49126-3
Online ISBN: 978-3-662-49127-0
eBook Packages: Springer Protocols