Abstract
In the past 20 years, the mRNA vaccine technology has evolved from the first proof of concept to the first licensed vaccine against emerging pandemics such as SARS-CoV-2. Two mRNA vaccines targeting SARS-CoV-2 have received emergency use authorization by US FDA, conditional marketing authorization by EMA, as well as multiple additional national regulatory authorities. The simple composition of an mRNA encoding the antigen formulated in a lipid nanoparticle enables a fast adaptation to new emerging pathogens. This can speed up vaccine development in pandemics from antigen and sequence selection to clinical trial to only a few months. mRNA vaccines are well tolerated and efficacious in animal models for multiple pathogens and will further contribute to the development of vaccines for other unaddressed diseases. Here, we give an overview of the mRNA vaccine design and factors for further optimization of this new promising technology and discuss current knowledge on the mode of action of mRNA vaccines interacting with the innate and adaptive immune system.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
1 Introduction
Vaccines provide the only durable protection against primary infections by pathogens. Since the smallpox vaccine development in 1798, countless lives and billions in health care costs have been saved (Plotkin 2014; Ozawa 2017). The World Health Organization (WHO) estimates that 2–3 million human lives are saved every year due to vaccination programs. Morbidity or crippling is prevented in numerous additional cases. Protective vaccines reduced annual poliomyelitis cases from 350,000 in 1988 to 33 in 2018. As of today, 26 infectious diseases can be prevented by vaccination, and four viruses have been eradicated from global circulation. Smallpox was eradicated in 1980 (World Health Organization 1980), wild type polio virus 2 and 3 in 2015 and 2019, respectively (https://www.who.int/news-room/feature-stories/detail/two-out-of-three-wild-poliovirus-strains-eradicated), and the animal pathogen rinderpest virus in 2011 (Mariner 2012). No other medical intervention is able to eradicate a disease.
Although a very successful medical intervention, existing vaccine technologies have their limitations, and progress in vaccine development is slowing down. Hence, new technologies are used to develop vaccines against pathogens such as SARS-CoV-2, which threaten our way of living.
In this review article, we summarize prophylactic vaccines with focus on mRNA-based vaccine technologies and their mode of action.
1.1 Established Vaccination Approaches
The main principle of vaccination is the induction of durable immunity against a pathogen by introducing either a part of the pathogen or the inactive or attenuated version of the pathogen into a vaccinee. The subsequent activation of the immune system, the induction of an adaptive immune response, and the establishment of a memory response against the pathogen allow the immune system to respond faster and more efficiently against this pathogen during subsequent infection to prevent disease manifestation. There are different classes of vaccines established. The first vaccines were attenuated versions of the pathogen that mimicked natural infection without causing disease in humans. These so-called live attenuated vaccines are able to replicate and express a variety of antigens. The resulting immune response is strong, broad, and long-lasting, sometimes due to low level of replication in the vaccinee. However, the attenuated virus might mutate and regain its pathogenicity, which occurred with the polio vaccine resulting in vaccine-associated paralytic poliomyelitis (Burns et al. 2014). Additionally, attenuated vaccines are reduced in their pathogenicity, but often cannot be safely administered to specific immunocompromised target populations, e.g., pregnant women, immunocompromised or human immunodeficiency virus (HIV) infected individuals (Hesseling 2009). Although the safety of these vaccines is excellent in most cases, the class has some limitations.
These vaccine-associated risks can be limited by using inactivated bacteria or viruses, e.g., the rabies, influenza, or the Hepatitis A virus vaccines (Plotkin 2014; Innis 1994). While inactivated pathogens are considered safer than live attenuated pathogens, they are also less immunogenic. This can partially be overcome by the use of adjuvants. The proper inactivation is key to safety, since incomplete viral inactivation might lead to vaccine-induced infections. Therefore, proof of inactivation is a critical release parameter for inactivated vaccines. The last severe cases associated with incomplete inactivation happened in 1955, when insufficient formalin-inactivation of the newly (inactivated) polio vaccine produced by Cutter pharmaceutical company caused 250 cases of atypical paralytic polio (Juskewitch et al. 2010). Today’s safety regulations and quality controls reflect lessons learned from those events and are designed to prevent reoccurrence. Nevertheless, quality inactivation control assessment can be demanding and intense. For the polio vaccine, WHO recommends a three week cell culture period with the vaccine virus (Chumakov et al. 2002); for rabies vaccine, it is even more intense, and it includes cell culture cultivation of the inactivated virus stained directly for virus replication or injected intracerebrally in mice (Bourhy 2007).
For live attenuated or inactivated whole organisms, replicating pathogens have to be produced in large quantities, often requiring individualized growth conditions for each vaccine, e.g., embryonated chicken eggs or cell culture for influenza virus. Reproducible vaccine production quality is challenging, and some vaccines suffer from high rate of batch failures. Moreover, the vaccine is more vulnerable to mutations that can decrease its efficiency. This is a problem especially for influenza vaccines. For egg-based influenza vaccines, the virus regularly adapts to the chicken cells by accumulating mutations within the receptor binding site which negatively influences vaccine efficiency, as observed for the vaccines of the last seasons (Zost 2017; Skowronski 2014).
Subunit vaccines, which contain only a protein of the respective pathogen, such as surface proteins (e.g., hepatitis B virus surface protein), or toxoids (e.g., Tetanus toxoid) are likely the safest. However, due to their high purity, they are less immune-stimulatory. They usually require an adjuvant, e.g., aluminum salts, which stimulate the immune system to support the induction of a protective immune response, but can induce adverse effects of their own (Petrovsky 2015). The first subunit vaccines were purified from cultured organisms, but with the rise of gene technology, recombinant proteins have become the standard. Manufacturing is more consistent and not as vulnerable to mutations as whole virus vaccines. However, for some pathogens, it is difficult to produce a stable, soluble antigen in the natural conformation needed to induce a protective immune response. Surface proteins, like viral envelope proteins, often have transmembrane domains and assemble into multimers. For the expression of such recombinant proteins, the introduction of stabilizing mutations or protein-engineering is necessary to produce the antigen in its natural conformation. This is exemplified by the HIV envelope (ENV) protein that is a trimeric transmembrane glycoprotein described as very unstable even during natural infection (Burton and Hangartner 2016). For recombinant protein expression, the full-length protein (160kD) is truncated to create a soluble gp140 protein, an internal protease cleavage site needs to be altered, and disulfide bonds are introduced to stabilize the trimer. Still, even small mutations can have a big impact on stability and immunogenicity (Beddows 2006; Sanders and Moore 2017). Similar results were reported for the respiratory syncytial virus fusion protein that is meta-stable, but a much better immunogenic in the pre-fusion conformation (Rossey et al. 2018).
Overall, prospective vaccines need to be easily manufactured, safe, and immunogenic. New vaccine technologies, such as viral vectors, DNA, and mRNA vaccines, have been developed showing promising features (Rauch et al. 2018).
1.2 Novel Vaccination Approaches
Viral vectors are engineered viruses, e.g., adenoviruses, adeno-associated viruses, or vesicular stomatitis viruses that encode a heterologous antigen. They are replication-deficient and deliver the antigenic sequence information into the host cell, which produces the antigen and presents it to immune system. Viral vectors allow a strong and diverse immune reaction to an antigen. At the same time, pre-existing anti-vector immune responses to the natural virus, e.g., adenovirus 5, can drastically decrease vaccination efficiency (Lemckert 2005).
DNA vaccines also deliver the antigenic sequence into the cell and induce transient antigen expression. The introduction of DNA into the host cell is challenging, since it has to reach the cell nucleus, crossing two cellular membranes, in order to facilitate antigen expression. Furthermore, the delivery of foreign DNA into a host cell comes with a risk of integration into the host genome, which could lead to unwanted side effects, including oncogenesis, depending on the integration site (Lee et al. 2018).
Using messenger RNA (mRNA) as a vaccine is a fairly new approach although it has been known since the early 90s that mRNA can induce antigen expression upon immunization and the induction of antigen-specific cytotoxic T cells (Wolff 1990; Martinon 1993). In 2000, Hoerr et al. confirmed and extended the potential of mRNAs as vaccines. They showed that immunizations with mRNA can be at least as effective as DNA in inducing cellular and humoral immune responses, i.e., cytotoxic T cells and antigen-specific antibodies (Hoerr et al. 2000). mRNA vaccines allowed the expression of antigens by the host cells and the expression of transmembrane proteins and viral glycoproteins with a natural glycosylation profile. Compared to DNA vaccines, mRNA can be more easily delivered into the cell, since it only needs to reach the cytoplasm for translation. Consequently and due to the absence of a reverse-transcriptase that could copy the mRNA into DNA, there is no risk of integration into the host DNA genome. Overall, use of mRNA is associated with a lower risk profile, as its production does not require cultivation of pathogens or any infectious materials at any step of the process. Production of an mRNA vaccine only requires the genetic sequence information, today often available via online databases. Moreover, only a limited number of antigens are expressed and for a short period of time. For a long time, RNA was perceived as a very unstable molecule for use as a genetic vector. However, handling in an RNAse-free environment and the formulation of the mRNA molecules allow the production of stable mRNA vaccines (Stitz 2017). In this chapter, we describe the underlying technology of mRNA vaccines in detail and discuss the immune responses that can be induced by mRNA vaccination.
2 mRNA Technology
A classical cellular mRNA has the minimal structural requirement of a 5′ cap, the open reading frame (ORF) and a 3′ poly(A) tail to enable efficient translation of the encoded protein. Untranslated regions (UTR) with regulatory function before and after the ORF can improve mRNA properties. Synthetic mRNAs are modeled after cellular mRNAs. They contain the ORF of the antigen complemented by UTRs, a 5′ cap, and a 3′ poly(A) tail (Fig. 1a). Synthetic mRNA vaccine are produced in a similar way (Schlake et al. 2012). First, the mRNA sequence is cloned into a plasmid downstream of a bacteriophage promotor, e.g., T7 or Sp6. The plasmid is subsequently linearized and used as a template for in vitro transcription by an RNA polymerase. After purification, the produced mRNA is formulated with proteins and/or lipids, which facilitate uptake by host cells and protect the mRNA against RNAses (Geall 2012).
Schematic structure of the mRNA and cap. a The general structure of an mRNA is based on a 5ʹ Cap, a 5′ UTR, an open reading frame (ORF) coding for the respective antigen, a 3′ UTR and a 3′ end containing a poly(A) stretch. b The 5′-cap structure is a N7-methylguanosine (methyl = purple) binding to the first nucleotide of the mRNA by 5′-5′ phosphodiester bond. Cap analogs can be modified at several position. P1 (green) and P2 (yellow) are used for methylation to generate the anti-reverse cap analog (ARCA). The phosphate chain (orange) can be prolonged or substituted with sulfur or other elements. At position P3 (blue), the next nucleoside will be attached by normal 5′-3′ bond, and Position P4 (pink) can be methylated to generate a cap1 structure
In the following paragraphs, different designs for these structural elements are presented, and their impact on mRNA stability and protein expression are reviewed.
2.1 5′ Cap Structure
Each eukaryotic mRNA starts with a 5′-cap structure. The most common natural cap is a N7-methylguanosine (m7G) which is connected to the mRNA via a 5′-5′ phosphodiester bond, followed by a ribose 2′-O-methylation on the first nucleotide (Banerjee 1980) (Fig. 1b). The cap interacts with cellular cap binding proteins, e.g., eukaryotic initiation factor 4E(eIF4E), which regulate mRNA processing, nuclear export, translation initiation, and prevent mRNA decay by blocking access of RNA decapping proteins, e.g., decapping protein 1 and 2 (DCP1/2). The cap is also involved in the discrimination between self and non-self mRNAs by the innate immune system (Lässig and Hopfner 2017; Galloway and Cowling 2019). To achieve maximum efficiency, synthetic mRNAs need to be capped, usually in parallel to, or subsequently to in vitro transcription (IVT).
There are three types of cap structures, cap0, where only m7G is added to the mRNA (m7GpppN), cap1, containing both m7G and 2′-O-methylation of the first nucleotide (m7GpppNm), and cap2, where m7G is followed by two methylated nucleotides (m7GpppNmNm). Cap1 and in theory cap2 are not only more efficiently incorporated into the mRNA and increase its translation, but they are also less likely to be detected by innate immune receptors. A detailed description of how mRNA vaccines interact with the innate immune response can be found in a latter part of this chapter (Sect. 3.2).
The first synthetic cap was mCap, a guanine dinucleotide m7GpppG and cap0 structure (Pasquinelli et al. 1995). It is incorporated co-transcriptionally into the mRNA by the RNA polymerase itself, which uses the mCap to initiate the IVT. However, T7 and other bacteriophage RNA polymerases can initiate at both guanines and therefore can incorporate mCap in forward (m7GpppG-mRNA) and reverse (Gpppm7G-mRNA) orientation with the approximated ratio of 30–50% reverse orientation (Pasquinelli et al. 1995). Reverse mCap is not recognized by the translation machinery, and no protein is expressed from these mRNA molecules. Hence, a significant portion of the mRNA will not be expressed.
There have been major efforts to improve capping of synthetic mRNAs (Table 1). Substitutions of the hydroxyl group at position C2 or C3 of m7G by a simple hydrogen (m7 2′dGpppG, m73′dGpppG) or the addition of a methyl group (m27 2′OGpppG, m27 3′OGpppG) prevent reverse incorporation of the cap and improve translation efficiency (Stepinski et al. 2001; Jemielity 2003). This cap was named “anti-reverse cap analog” (ARCA) (Stepinski et al. 2001). Later, mostly the m7 2′OGpppG is referred as ARCA. Additional modifications increase the translation efficiency further and are summarized in Table 1. They include the extensions of the phosphate chain, while a tetraphosphate (m7GppppG) increases translation efficiency due to higher binding efficiency to eIF4E, a pentaphosphate (m7GpppppG) shows decreased translation efficiency by preventing eIF4E release (Jemielity 2003). The insertion of bridging modifications, e.g., dichloromethylene insertions (m7Gpp-CCl2-ppG) or sulfur substitutions (m7 2′O GppspG, named β-S-ARCA), prevents decapping (Rydzik 2017; Grudzien-Nogalska et al. 2007). The β-S-ARCA showed a nearly doubled half-life in vitro and improved stability in primary dendritic cells (DC) (Kuhn 2010). Furthermore, it induces increased T cell responses in vivo (intranodal application of unformulated mRNA) compared to the regular ARCA (Kuhn 2010). Other modifications affect the m7G and can improve the mRNA stability as well. A benzyl at position P2 (Fig. 1b) enhances overall translation efficiency due to improved eIF4E binding, although it might be more sensitive to decapping by Dcp1/Dcp2 (Kocmik 2018). Finally, the locked nucleic acid with a modification of the first guanosine has a lower capping efficiency compared to ARCA, but still a higher translation efficiency due to improved binding to eIF4E (Kore et al. 2009).
More recently, TriLink has developed a synthetic cap1 (m7GpppNmN), called CleanCap®, that is added co-transcriptionally (Vaidyanathan 2018; www.cleancapmrna.com). According to the company, it outcompetes ARCA (cap0) in capping efficiency (95 vs. 70% for ARCA) and translation efficiency. The CleanCap® is available as a natural m7GpppNmN cap or with the ARCA modification (m7 3′dGpppNmN) and both with the different variants of the second and third nucleotide NN = GG, AU or AG.
Alternatively, caps can be added post-transcriptionally by the vaccinia virus capping complex (Schnierle et al. 1992; Venkatesan et al. 1980; Meis et al. 2006), e.g., the commonly used synthetic cap structure, called ScriptCap. It is post-transcriptionally incorporated by subsequent incubation with a capping enzyme adding m7G, and a methyltransferase adding the 2′-O-methylation (Schnierle et al. 1992). Although the capping efficiency is nearly 100%, the addition of one or two enzymatic reactions and a purification step adds time and costs to the manufacturing process (Meis et al. 2006). Methyltransferases can also be used to add 2′-O-methylations to an existing cap0 to enhance translation efficiency (Richner 2017).
2.2 Untranslated Regions (UTR) of mRNA
UTRs are an essential part of most eukaryotic mRNAs and all RNA viruses. They contain regulatory elements that recruit cellular factors to the mRNA 5′ and 3′ ends and with further optimization can improve translation efficiency and mRNA stability (Ahmed et al. 2011).
mRNA translation is initiated by eIF4E initiation factor interaction with the cap and assembly of the initiation complex 43S (Ahmed et al. 2011). TISU (translation initiation of short 5’UTRs, GCCAGAaug) and Kozak (GCCRCCaugG) sequences are translation initiation elements that allow binding of the ribosome 43S initiation complex which scans the mRNA for the first AUG start codon (Elfakess et al.2011; Kozak 1991). Weak AUG context sequences around the start codon can be skipped by the ribosome and translation initiated at the next AUG, resulting in shorter or different proteins, a process called leaky scanning/AUG skipping (Kozak 2005). Even though the Kozak sequence alone is sufficient to induce translation of the mRNA, a longer untranslated region upstream of the start codon can lead to higher translation efficiencies (Kozak 1991).
In humans, 5′UTRs, the regions upstream of the start codon, have a median length of 218 bp (Leppek et al. 2018). They can enhance the translation efficiency, e.g., the 5′UTR of Hsp70, β-globin, and tobacco etch virus increase protein expression level even when cloned upstream of a heterologous ORF (Schlake et al. 2012; Kozak 1991; Vivinus 2001; Schlake et al. 2019; Holtkamp 2006). UTRs can also decrease or even prevent protein expression. The iron responsive element (IRE), naturally found in the ferritin and the iron transporter ferroportin mRNAs, is bound by iron-regulatory proteins in low iron conditions. While the IRE in the ferritin mRNA is at the 5′ UTR, the interaction prevents association of the mRNA with the ribosome, causes translation inhibition and degradation, to reduce ferritin expression and storage of iron under iron starvation condition. Ferroportin mRNAs have the IRE on the 3′ end, which has the exact opposite effect and increases the expression of the transporter to maintain iron levels in the cell (Ahmed et al. 2011; Leppek et al. 2018; Muckenthaler et al. 2017).
Thus, therapeutic mRNA translation can be improved by adding a particular 5′ UTR. Variety of secondary UTR structures can be formed depending on length, GC content, and sequence, affecting translation efficiency. 5′ UTRs with a high GC content are more likely to have a complex secondary structures (Leppek et al. 2018), for instance stem loops which can favor 43S ribosome recruitment through the transacting factor eIF3. For example, the 5′ UTR of the interferon γ (IFNγ) mRNA forms a pseudoknot. In turn, this dsRNA structure activates the innate immune response locally. The activation leads to translation arrest and represents a negative feedback loop to prevent uncontrolled IFNγ production (Ben-Asouli et al. 2002). Other secondary structures such as stem loops, IRE, hairpins, and RNA G-quadruplexes might have similar impact on the translation efficiency (Leppek et al. 2018).
Interestingly, Trepotec et al. described a highly efficient minimal 5′ UTR of only 7–8 nucleotides (Trepotec et al. 2018). In combination with a Kozak sequence or a TISU element, these short sequences increased protein levels over the gold-standard 5′ UTR α-globin (30 bases).
3′ UTRs are located downstream of the ORF. They are usually longer than 5′ UTRs (median length 1200 nucleotides) and regulate the stability of the mRNA (Mayr 2008; Jan et al. 2011). Like 5′ UTRs, 3′ UTR sequences contain motifs that recruit RNA binding proteins (RBP) which act as linkers between the mRNA and functional proteins. Structural elements such as IREs and AU-rich elements (ARE) can induce repression of translation, deadenylation (shortening of the poly-A tail), decapping, or cleavage and lead to mRNA decay (Mayr 2008; Koh et al. 2019). Sequence motifs found only in the 3′UTRs are seed regions for miRNAs, which may also promote mRNA decay (Ahmed et al. 2011; Mayr 2008; Jia et al. 2013; Rabani et al. 2017). Some 3′ UTRs, like those from α- and β-globin, can increase mRNA stability (Holtkamp 2006; Wang et al. 1999). The α-globin 3′ UTR recruits the α-complex, which stabilizes the poly(A) binding protein (PABP) to protect the poly(A) tail and prevent deadenylation (Wang et al. 1999; Bernstein et al. 1989). In tumor tissues, it has been observed that 3′ UTR shortening significantly increases oncogene mRNA half-life (Mayr 2008). Additionally, 3′ UTRs can contain zipcodes, which serve as motifs for RBPs that interact with tubulin-associated motors and transport mRNAs closer to where the protein is needed, e.g., the leading edge of actin filaments for β-actin mRNA (Lawrence and Singer 1986). 3′ UTRs can also affect the function of the translated protein by influencing the location of the mRNA and its translation impacting protein interactions (Mayr 2008). For example, it has been demonstrated for the mRNA encoding CD47, that only the long 3′ UTR mRNA can lead to a cell surface expressed CD47 and interaction with SET, while the short 3′ UTR mRNA lead to intracellular CD47 expression. Cell surface expressed CD47 can prevent the cell from phagocytosis, while intracellular CD47 is involved in the induction of apoptosis. Even though both proteins have the same amino acid sequence, the 3′ UTR defines their functionality (Berkovits and Mayr 2015). Hence, UTR regulatory elements are able to strongly impact protein function.
For synthetic mRNAs, selection of suitable 3′ UTR can prolong expression and protein level. The inclusion of multiple 3′ UTRs can also increase stability and translation efficiency. A screen for stable mRNA constructs in dendritic cells identified a combination of the 3′ UTRs of mtRNR1 and AES mRNAs that increased protein expression in vitro and in vivo and led to higher T cell responses upon immunization (Orlandini von Niessen 2019). Until very recently, limited information on UTR motifs and their effects on translation efficiency was available, and choice of UTRs had to be evaluated empirically. Improvements in sequencing technologies and machine learning have identified a plethora of novel 3′UTR motifs and allow for predicting the impact of UTR sequences on ribosome loading and translation in human cells (Rabani et al. 2017; Sample 2019). The technology supports generation of 5′UTR sequences with targeted ribosome loading and protein expression levels (Sample 2019). It is likely that selection of UTRs will increase in the future mRNA designs.
2.3 3′End of the mRNA
The poly(A) tail forms the very 3′ end of an mRNA and is of varying length (Stewart 2019). Its intracellular shortening by deadenylases regulates transcript half-life. The poly(A) is also involved in the initiation of translation via interaction of poly-A binding proteins with the translation initiation complex at the 5ʹ end (Ford et al. 1997). Cellular mRNAs are polyadenylated in the nucleus independently of template and are subsequently exported to the cytoplasm (Stewart 2019).
Synthetic mRNAs can be polyadenylated as part of the IVT by encoding a defined number of As on the plasmid template, or by a poly(A) polymerase that adds poly(A) tails of varying lengths (30–200 nts) to the transcript after the IVT (Beverly et al. 2018; Kuhn et al. 2011). The plasmid encoded poly(A) is of defined length varying only in a few nucleotides (Beverly et al. 2018). In general, at least 30 As are needed for a functional tail, but longer tails confer higher stability (Ford et al. 1997). Sixty As seem optimal to induce high translation efficiency in most cell lines (Elango et al. 2005). Longer poly(A) tails led to decrease in translation efficiency in this context. Some immune cells seem to be exceptions. Translation efficiency can increase to lengths of 120 As for dendritic cells, while the maximal efficiency was reached at 300 As for primary T cells (Holtkamp 2006; Ford et al. 1997; Elango et al. 2005; Grier 2016). However, long A-stretches are challenging to produce. In bacteria, the poly(A) tail encoded on the plasmid template is shortened to approximately 70 nt as a result of recombination during amplification (Grier 2016). This can be avoided by segmenting the poly(A) tail into pieces of 40–60 nt separated by a few non-A nucleotides, without affecting mRNA stability or translation efficiency (Trepotec et al. 2019). Non-A nucleotides are tolerated within stretches of As. However, translation efficiency was ~30% lower when dendritic cells were transfected with in vitro transcribed mRNA ending with five non-A nucleotides compared to a clean poly(A) tail (Holtkamp 2006). In primary T cells, up to 6 non-A nucleotides can be tolerated without pronounced reduction of translation efficiency if the extension contains exclusively GC (Grier 2016). An extension containing one or more Us strongly impaired the translation. This observation is especially relevant for the linearization of the template plasmid. Most commonly used restriction enzymes are of type II and generate a 3′end with 1–5 nucleotides at the end of the poly(A) (Elango et al. 2005). Recently, type IIS restriction enzyme has been employed for plasmid linearization to generate blunt A ends (Holtkamp 2006; Elango et al. 2005)
As an alternative to a poly(A) tail, a histone stem loop (HSL) can be used to terminate the mRNA. Histone mRNAs are usually not polyadenylated and end in a conserved 26 nt long stem loop structure that protects the transcript from degradation during the S-phase of mitosis (Kaygun and Marzluff 2005; Marzluff 1992). However, a few histone mRNAs are polyadenylated downstream of the stem loop in non-growing cells, allowing for better regulation of transcript levels. Thus, poly(A) and HSL are not mutually exclusive (Mannironi et al. 1989).
Synthetic mRNAs combining optimized 3′UTR sequences, poly(A) length, and a clean ending can improve transcript levels up to 100-fold, and peak protein levels and expression time fivefold (Holtkamp 2006).
2.4 ORF Optimization and Global Modifications
ORF sequences can have a big impact on mRNA half-life and translation. Each species has its own codon bias (Novoa and Ribas de Pouplana 2012), which can lead to increased protein expression up to 1000-fold (Gustafsson et al. 2004). The codon optimization of the synthetic mRNA ORF could strongly increase the expression of the respective encoded antigen. Moreover, the codon usage within the ORF may differ (Tuller and Zur 2015; Clarke and Clark 2010). Less structured 5′end of the ORF supports ribosome scanning and binding, while the rest of the ORF has been described to benefit from higher structure that slows down the ribosome and improves translation and protein folding (Tuller and Zur 2015; Mauger 2019; Ding et al. 2014). The effect observed on cellular mRNAs can be leveraged to improve synthetic mRNA vaccine technology in the future. Currently, most mRNA platforms use a rather simple GC enrichment (Rauch et al. 2018).
Besides the optimization of the sequence itself, the nucleotides can be modified as well. In vitro transcribed (IVT) mRNAs are usually based on the classical natural nucleotides (A, T, G, C). Unfortunately, delivered ssRNAs are recognized as immunogenic and activate different antiviral innate immune mechanisms leading to cytokine release, translation breakdown, and RNA degradation. Even though an innate immune activation is necessary for a vaccine to induce an effective adaptive immune response (explained in more details in the “mode-of-action” section), a too strong innate immune reaction can be detrimental by interfering with the translation efficiency, impairing antigen expression, and as such inhibit the induction of an adaptive immune response.
A comparison of different host RNA species to IVT RNA or bacterial RNA showed that tRNA and rRNA, which contain a high amount of chemically modified nucleotides, are much less immune stimulatory (Karikó et al. 2005). Subsequently, several modified nucleotides have been analyzed for optimizing translation efficiency and reducing innate immune reaction. In this review, we focus on the five most commonly used modified nucleotides: pseudouridine [ψ, the most abundant nucleotide modification in host cell RNAs (Spenkuch et al. 2014)], N1-methyl-pseudouridine (m1ψ), 5-methylcytidin (5mC), N6-methyladenosine (m6A), and 2-thiouridine (s2U). The modified nucleotides have three major effects: reduction of innate immune stimulation, reduction of mRNA impurities, and effect on protein expression levels.
All modified nucleotides reduce activation of pattern recognition receptors (TRLs; RIG-I, and MDA) to prevent extensive cytokine release. In dendritic cells, m6A and s2U completely abolish the TLR activation (TLR 3, 7 and 8), while ψ and m5C only slightly reduce TLR activation and reach full suppression only in combination (Karikó et al. 2005; Andries et al. 2015). Subsequently, it has been demonstrated that RIG-I signaling is impacted by modified nucleotides. m6A completely prevents interaction with RIG-I (Durbin et al. 2016). In contrast, other modification such as ψ, m1ψ, and 5mC RNAs bind to RIG-I, but inhibit signaling pathway activation by preventing the conformational change of RIG-I (Durbin et al. 2016).
In addition to their effect on innate immune receptors, modified nucleotides can influence the IVT reaction. Antisense RNA is a common side product of IVT. It creates dsRNA region that can activate MDA5, which usually detects long dsRNA (Kato 2008). RNA impurities are usually removed by HPLC purification (Karikó et al. 2011). Interestingly, incorporation of ψ, m1ψ, or 5mC (but not m6A) into synthetic mRNAs reduces the occurrence of such unintended products (Mu et al. Jun. 2018; Baiersdörfer 2019).
The inhibition of immune cell activation and cytokine release could be confirmed in several mouse studies after application of IVT RNA i.v. (Karikó 2008; Kormann 2011; Tusup et al. 2018), i.d., i.m. (Andries et al. 2015) or into the lung (Andries 2013). A systemic cytokine release of IFNα, IFNβ, TNF, and IL-6 was inhibited (Karikó 2008). Only few studies could not detect a difference between RNA with natural nucleotides or ψ-containing RNA (Kauffman 2016; Thess 2015).
Beside the evasion of immune activation, modified nucleotides can influence mRNA stability and translation efficiency. Because of the strong decrease of translation efficiency, m6A and s2U are not suitable modification for mRNA vaccines, (Karikó et al. 2005; Karikó 2008). Effects on translation efficiency by ψ, m1ψ, or 5mC containing mRNAs are debatable and seem to be cell type dependent. Ψ and 5mC RNAs have been described to increase translation efficiency in DCs, PBMCs, and MEFs (Karikó et al. 2011; Karikó 2008). On the other hand, ψ-containing mRNAs have the same or even lower translation efficiency compared to natural nucleotide mRNAs in macrophages, HeLa cells, and keratinocytes (Andries et al. 2015; Kauffman 2016; Thess 2015; Uchida et al. 2015; Loomis 2018). In HEK293 cells, reported results differ, probably depending on delivery methods and further differences in the RNAs (Karikó et al. 2011; Tusup et al. 2018; Svitkin et al. 2017). mRNA secondary structure influences protein expression level as well. Mauger and colleagues observed that certain modified nucleotides, especially m1Ψ, can stabilize the secondary structure to increase translation efficiency, whereas mo5U destabilizes RNA structures (Mauger 2019). m6A destabilizes secondary structures such as hair pins (Kierzek and Kierzek 2003). Modified nucleotides can prevent the activation of intracellular antiviral defense mechanism, like PKR-induced phosphorylation of eIF2α, leading to a general translation inhibition and OAS-induced RNase L-mediated RNA degradation. Consequently, the inhibition of PKR and OAS ensures RNA stability and translation upon intracellular delivery (Svitkin et al. 2017; Anderson et al. 2011; Anderson 2010).
In vivo, most studies confirm a higher and prolonged protein expression by nucleotide-modified mRNAs (Baiersdörfer 2019; Kormann 2011; Karikó 2008; Tusup et al. 2018; Anderson et al. 2011; Karikó et al. May 2012). Andries et al. tested the three most potent modified nucleotides in mice by i.d. and i.m. application (most relevant for vaccines) and showed that m1ψ outperforms ψ and 5mC by 13-fold in protein translation in vivo (Andries et al. 2015). In line with this observation, Drew Weisman and colleagues developed several m1ψ-containing mRNA vaccines against zika, HIV, and influenza (Pardi et al. 2017, 2018, 2019).
However, two studies could not confirm any improvements of modified nucleotides (ψ) over natural nucleotides in terms of cytokine induction and translation efficiency (Kauffman 2016; Thess 2015). Importantly, the mRNA used by Thess et al. does not have a traditional poly(A) 3′end but rather ends with a histone stem loop (Thess 2015). The secondary structure, and hence the function of this mRNA element, is potentially affected by incorporation of ψ, explaining the observed differences.
In most studies, uridine or cytosine has been replaced completely by the modified nucleotides (= 100% modification). However, based on the observation of host RNA species, where only a fraction of the nucleotides are modified, Kariko et al. observed that incorporation of only 10% modified nucleotides (ψ, 5mC, or m6A) is sufficient to reduce TNF release by 50% in DCs (Karikó et al. 2005). For complete inhibition, at least 90% of the nucleotides had to be modified. Similar results have been observed in RAW cells (macrophage cell line) (Uchida et al. 2015).
In summary, all properties of a synthetic mRNA can affect the mRNA efficiency and quality, including UTRs or choice of nucleotides.
3 Mode of Action of mRNA Vaccines
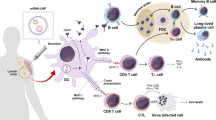
The general principle of any vaccine is the durable induction of a protective immune response against an antigen. For mRNA vaccines, this is achieved by delivering the antigenic sequence into the vaccinee’s cells, so that they can express the encoded protein and present it to the immune system. mRNA vaccines have several advantages over recombinant protein and whole viral particle vaccines: ssRNA itself has an adjuvant-effect, which abolishes the need of an additional adjuvant (Edwards 2017). Furthermore, antigen expression by the host cells facilitates the correct folding of the antigen and enables the incorporation of transmembrane proteins presented on the cell surface in their native conformation. Viral antigens are expressed similarly during a viral infection. Like other vaccines, mRNA vaccines can be administered by different routes. For practical reasons, most vaccines are given intramuscularly (i.m.), even though various clinical trials suggest that intradermal (i.d.) administration induces stronger immune responses or needs less vaccine to induce the same response compared to i.m. or s.c. (subcutaneous) immunizations (Hickling et al. 2011; Zhang et al. 2015). It is believed that more professional antigen presenting cells (APC), such as dendritic cells (DCs), reside in the skin, and thus induce a stronger immune response. APCs take up the mRNA cargo, which stimulates innate immune signaling, and in response the cells migrate to the draining lymph node. There, the APCs present the antigen to B- and T cells and activate the adaptive immune response. We will discuss each of these steps in more detail in the following paragraphs.
3.1 Innate Immune Sensors Detecting mRNAs
Mammalian cells detect a variety of pathogen-associated molecular patterns (PAMPs) with the help of pattern recognition receptors (PRRs). Some PAMPs are molecules that only exist on pathogens, e.g., bacterial lipopolysaccharide or flagellin (Mogensen 2009). Others exist in the host (self) and the pathogen (non-self) and need to be distinguished by PRRs, e.g., unmethylated DNA of bacteria.
RNA as a PAMP is identified based on its location and structure. For example, mRNAs have no function outside of the cells. If cells take up extracellular RNA species via endocytosis, they are detected as non-self by PRRs in the endosome, specifically toll-like receptors (TLRs) 3, 7, and 8 (Georg and Sander 2019). TLR3 binds to dsRNA and activates TRIF (TIR-domain-containing adapter-inducing interferon-β), inducing a signaling cascade that leads to IRF3 activation, and the expression of type I interferons and IP-10. Both TLR7 and 8 sense ssRNA and signal through MyD88 and IRF7, but they show a cell type specific expression pattern (Patinote 2020). TLR7 is only expressed in plasmacytoid dendritic cells (pDCs) and B cells, where its activation leads to type I interferon production. TLR8, on the other hand, is found in monocytes and monocyte-derived DCs (mDCs) and induces TNF, IL-6, IL-12, and MIP-1α.
The cytoplasm contains a different set of PRRs, which have to distinguish non-self RNA from self RNA, especially mRNA. Retinoic acid-inducible gene (RIG)-I and melanoma differentiation-associated proteins 5 (MDA5) sense double-strand RNA (dsRNA) (Georg and Sander 2019; Brisse and Ly 2019). RIG-I prefers blunt end dsRNA ligands of at least 13 bp and with 5′-PPP—or Cap0-ends, to which it binds and oligomerizes (Devarkar 2016; Schmidt 2009). In contrast, MDA5 binds to dsRNA of at least 2000 bp, such as viral genomes, to which it binds at any position and starts polymerizing as well. RIG-I/MDA5 filaments then recruit and activate mitochondrial antiviral signaling protein (MAVS). MAVS activation leads to induction of IRF3 and IRF7 signaling, and expression of NF-κB and type I interferon. Furthermore, cytoplasmic NOD-like receptors like NRLP3 can also respond to dsRNA and to a lesser extent to ssRNA (Allen 2009). This induces the formation of the inflammasome. It is a major mechanism in macrophages and leads to a strong release of IL-1β and induction of apoptosis (Rajan et al. 2010).
Other dsRNA sensors like protein kinase RNA-induced (PKR) and 2′-5′-oligoadenylatesynthetase (OAS) are IFN-induced. In turn, they induce global translation arrest and RNA degradation upon activation. PKR is also an intermediary of TLR3 signaling and can stimulate the expression of IL-1β and IL-18.
Distinguishing self from non-self single-stranded RNA (ssRNA) in the cytoplasm depends as well on the mRNA cap structure. IFN-inducible proteins with tetratricopeptides (IFITs) detect ssRNAs and discriminate them based on their 5′ structure. IFIT1, for example, detects a missing 2′–O–methylation of the first nucleotide (i.e., cap0) and sequesters 5′-PPP-mRNAs (Fensterl and Sen 2015). Upon recognition of their ligand, IFITs associate with the RNA and prevent its translation or replication. As their name indicates, IFITs are induced by IFN signaling and are not constitutively expressed.
Downstream of all PRRs is the activation of antimicrobial signaling pathways and the expression and secretion of type I interferons, i.e., IFN-α and IFN-β. They can bind to their receptors (IFNAR) found on all cells and subsequently can induce paracrine activation of the “antiviral state” of neighboring cells through the expression of IFN-stimulated genes (ISGs). ISGs include PKR and OAS, which broadly inhibit protein translation. For example, PKR phosphorylates translation factor eIF2α in the presence of dsRNA, leading to a general inhibition of protein synthesis (Anderson 2010). OAS signaling leads to activation of RNase L, which degrades RNA (Anderson et al. 2011). Additional ISGs, like myxovirus resistance 1 (MX1) or apolipoprotein B mRNA-editing enzyme catalytic polypeptide (APOBEC1), have more specific antiviral functions (Sadler and Williams 2008). More importantly, IFNα/β activate immature DCs and turn them into APCs by inducing the expression of i) MHC molecules, ii) co-stimulatory proteins, e.g., CD80 and CD86, iii) chemokine receptors, and iv) cytokines such as IL-12 (Kranz et al. 2016). This allows DC to present more antigens, better activate T cells, migrate to the lymph node where the T and B cells are located, and influence the adaptive immune response according to the antigen, i.e., IL-12 production drives TH-1 differentiation (McNab et al. 2015). Other cytokines downstream of innate immune sensors includes pro-inflammatory cytokines, TNF, IL-1α/β, IL-6, and IL-8. Therefore, the early induction of type I interferons and cytokines is crucial for mounting a strong, long-lasting adaptive immune response against a pathogen.
3.2 Immune Activation by mRNA Vaccines (Adjuvancy vs. Overactivation)
Messenger RNA vaccines reliably stimulate the innate immune response independently of the encoded antigen. Consequently, the innate response induces a strong adaptive immune response. Thus, unlike subunit vaccines, mRNA vaccines do not require an adjuvant for immune stimulation but are innate stimulatory as such (Edwards 2017; Beverley 2002). After administration, cells at the injection site take up the mRNA vaccine. Most mRNA vaccines used today are formulated as lipid nanoparticles (LNP), which are taken up by endocytosis, and after endosomal escape, reach the cytosol where the mRNA is expressed (Edwards 2017; Kranz et al. 2016; Devoldere et al. 2016). Some of these cells will be APCs, especially dendritic cells (DCs), and to a certain degree, macrophages. They detect the non-self mRNA in the endosome via TLR7/8, mount an innate immune response, and produce pro-inflammatory cytokines (Kranz et al. 2016). Several studies confirm that TLR7 activation is predominantly responsible for the acute inflammatory response after mRNA vaccination (Kranz et al. 2016; Fotin-Mleczek 2011). Conversely, the T cell response is strongly impaired in TLR7–/– mice after vaccination with mRNA vaccines, showing the importance of TLR7 activation and subsequent cytokine release for T cell response (Kranz et al. 2016). When naked mRNA is delivered by electroporation, PRRs in the cytoplasm might be activated (Devoldere et al. 2016; Iavarone et al. 2017).
A broad range of pro-inflammatory cytokines has been detected at the injection site upon i.m. or i.d. injection, including the acute phase cytokines IL-6, IL-1β and TNF, leading to DC activation and migration, lymphocyte activation, and increased antibody production (Edwards 2017; Lutz 2017; Kowalczyk 2016). Additional cytokines and chemokines have been detected in the muscle after i.m. injection (Lutz 2017). MIP-1α, MIP-1β, CXCL9, MCP-1, and CXCL1 recruit more APCs, as well as NK-, B-, and T cells (Carr et al. 1994; Wolpe and Cerami 1989; Groom and Luster 2011; Tannenbaum et al. 1998). Dendritic cells, and to a certain degree macrophages, are the main actors in the cytokine production (Kranz et al. 2016). Overall, there is a local inflammatory reaction. However, the cytokine release has been reported to be short lived, peaking between 6 and 14 h post immunization and usually returning to baseline after 24 h (Edwards 2017; Lutz 2017; Kowalczyk 2016; Broos 2016). Generally, no or very low systemic cytokine release could be observed in comparison to the local reaction (Lutz 2017; Kowalczyk 2016).
Within 4 h of after immunization, high antigen levels are expressed in DCs and monocytes (Broos 2016; Liang 2017). Uptake and mRNA expression are concurrent with upregulation of activation markers, such as CD40, CD80, and CD86 as well as MHC for efficient antigen presentation (Kranz et al. 2016, Edwards 2017; Broos 2016; Liang 2017; Scheel 2004). Activated DCs have also been shown to migrate to the draining lymph nodes (dLN) and create a pro-inflammatory milieu (Kowalczyk 2016; Carralot 2004). After i.d. injection of protamine-formulated mRNA, increased chemokine (CXCL9 (MIG), CCL2 (MCP1), CCL3 (MPI-1α), CCL4 (MIP-1β)), and cytokine (TNFα, IFNα, IFNγ, IL-1) levels have been found in the dLN (Edwards 2017; Kowalczyk 2016). This results in recruitment and activation of lymphocytes and the induction of adaptive immune responses (Carr et al. 1994; Wolpe and Cerami 1989; Tannenbaum et al. 1998).
Taken together, i.m. or i.d. injections of mRNA vaccines induce a pro-inflammatory environment in the draining lymph nodes within 24 h, enabling DCs to present the antigen to the adaptive immune system and creating highly immune-stimulatory conditions to develop a strong immune response against the respective encoded antigen.
Unfortunately, due to the mRNA production process, incompletely capped or partially degraded mRNAs, as well as non-specific oligoribonucleotides, may appear as side products of RNA polymerase in in vitro transcription, can be contained in the vaccine, and can create unintended RNA PAMPs (Karikó et al. 2011). These would be detected by additional PRRs, result in an overstimulation of the innate immune system and an excessive inflammatory reaction, which can be detrimental to the adaptive immune response. The important role for type I interferons in connecting innate and adaptive immune systems has been described. IFNα is necessary for proper DC activation and maturation, leading to increased expression of co-stimulatory molecules (Kranz et al. 2016). Furthermore, it increases antigen cross-presentation and induces the release of chemokines for T cell recruitment (Crouse et al. 2015). The role of IFNα on the immune response upon mRNA vaccination seems to be double-edged. High levels of IFNα are associated with various symptoms of malaise. Prolonged IFNα expression has been linked to development of autoimmune disease in a different context (Wills et al. 1984; Rönnblom 1991; Borg and Isenberg 2007). Locally, it upregulates antiviral effectors like OAS and PKR, potentially degrading the mRNA vaccine and preventing its translation and antigen presentation (Devoldere et al. 2016). Along those lines, a higher antigen expression was observed in IFNAR–/–DC after mRNA/lipoplex delivery (Pollard 2013). It is unknown to what extent a reduced antigen expression might affect vaccine efficacy. mRNA vaccine that is not expressed, or only at low levels, will not be able to induce an efficient adaptive immune response. In general, the role of IFNα and its effects on immune responses are incompletely understood (McNab et al. 2015). Depending on the timing, level, and environment of IFNα expression, it might stimulate or dampen T cell responses and regulate antigen expression in different ways (Kranz et al. 2016; Crouse et al. 2015; Pollard 2013; Beuckelaer 2016; Beuckelaer et al. 2017). Kranz et al. reported that IFNα induction is important for CD8+ T cell effector function (Kranz et al. 2016). Intravenous immunization of IFNRA–/–mice with mRNA/lipoplex showed impaired and shortened DC activation and significantly lower levels of IFNα. Antigen-specific CD8+ T cells were induced, but had a strongly impaired effector function lower Granzyme B, IFNγ and TNF expression. On the other hand, De Beucklear et al. claimed that the application of mRNA/lipoplex subcutaneously (s.c.) or i.d. in IFNAR–/–mice showed a much stronger cytotoxic T cell (CTL) response than in WT mice (Beuckelaer 2016). In a cytotoxicity assay, only a small portion of the antigen presenting target cells (peptide pulsed) could be eliminated by antigen-specific CTLs from WT mice, while there was a complete eradication of target cell by antigen-specific CTLs from IFNAR–/–mice. Based on these opposing observations, De Beucklear hypothesized that kinetics of IFNα and TCR signaling determine the positive or negative effect of IFNα release (Beuckelaer et al. 2017). When IFNα release and TCR stimulation occur simultaneously, like after i.v. injection, T cells are strongly stimulated, and IFNα serves as the third signal leading to differentiation and effector function (Kranz et al. 2016; Broos 2016; Le Bon et al. 2014). However, if the IFNα signal is induced before TCR stimulation, it might have the opposite effect leading to anergy and cell death/inhibition of proliferation. The hypothesis explaining these opposing IFNα effects is based on the ability of IFNAR to signal either through STAT4 (positive, pro-inflammatory, proliferative) or STAT1 (anti-proliferative, apoptotic) and is reviewed by Crouse et al. (2015). Upon TCR activation, T cells upregulate STAT4 while maintaining STAT1 level constantly low.
In conclusion, a balanced pro-inflammatory innate immune response is supportive of a subsequent adaptive immune response. The exact factors needed for optimal immune response to the mRNA vaccines, and especially the role of IFNα are subject to further studies. It is assumed that a potent but local (injection side and draining LN) pro-inflammatory environment is beneficial to the induction of both, a strong humoral and cellular immune response. Importantly, a substantial systemic cytokine release is not preferred, since it might be associated with side effects like fever, headache, chills, and fatigue induced by acute phase cytokine and type I IFNs. These reactions have been reported after mRNA vaccination in clinical phase 1/2 studies, including studies using modified and non-modified nucleotides (Gruys et al. 2005; Alberer 2017; Mulligan et al. 2020; Jackson et al. 2020; Bahl et al. 2017). Moreover, cytokine release might lead to a decline of pDCs by the intrinsic apoptosis (Swiecki et al. 2011). Most mRNA platforms are describing only a transient local cytokine release without systemic inflammation (Lutz 2017; Pardi et al. 2017).
3.3 Adaptive Immune Response
Depending on the pathogen, different immune effectors are likely to confer protection. Most acute viral infections will be prevented by neutralizing antibodies as described for influenza, rabies, or measles viruses (Nigg and Walker 2009; Haralambieva et al. 2019; Padilla-Quirarte et al. 2019). RNA viruses with high antigen variability can rapidly escape neutralizing antibodies by mutations in surface antigens. In this case, a strong cellular immune response can limit the disease by killing cells actively replicating the virus, stopping viral spread to reduce pathology. Some therapeutic vaccines for HIV or other chronic viral infections aim to boost the cytotoxic T cell response to control viral replication (Ndhlovu 2015). For mRNA vaccines, both have been described, the induction of protective humoral as well as cellular immune responses.
3.3.1 Humoral Immunity
Indeed, mRNA vaccines induce strong antibody responses to a variety of viral infections, e.g., influenza (Pardi et al. 2018; Lindgren 2017; Petsch 2012), rabies (Lutz 2017), and HIV (Pardi et al. 2019). A nucleotide-modified mRNA/LNP vaccine encoding influenza virus hemagglutinin outperformed the inactivated influenza virus (IIV) vaccines and live attenuated virus vaccines in a mouse model (Pardi et al. 2018). The hemagglutination inhibition (HAI)-titer, a surrogate marker for neutralizing antibodies against influenza, was 40-times higher in mRNA vaccinated animals than in IIV-vaccinated mice after two i.d. immunizations and remained stable for at least 13 months. Similarly, a protamine-formulated mRNA also induced a strong antibody response with HAI-titers above the correlate of protection (1:40) even after a single vaccine dose (Petsch 2012). In a passive serum transfer experiment with a subsequent viral challenge, it was demonstrated that the induced antibodies are sufficient to confer full protection in mice. Impressively, this vaccine was not only efficient in adult mice, but could induce protective immunity in newborn mice and very old (18 month) mice (Petsch 2012). Similar results were obtained in newborn piglets for an mRNA vaccine encoding the rabies G protein (Schnee 2016). These findings show that mRNA vaccines might be efficacious in populations at increased risk of sever outcome such as newborns and aging individuals. Follow-up studies in non-human primates (NHP) using LNP-formulated mRNA confirmed the potency of mRNA vaccines to induce neutralizing antibody titers at least as high as licensed vaccines, e.g., influenza mRNA vaccine vs. Fluad, or rabies mRNA vaccine versus licensed inactivated Rabies vaccine (Lutz 2017). Functional antibody titers in terms of HAI- titers in NHPs were stable for at least 1 year. The application route (i.m. vs. i.d.) does not seem to make a significant difference in this model. However, prime i.d. injections of modified mRNA vaccines induce a faster increase in antibody response (Lindgren 2017). Both immunization routes reach the same antibody titer plateau and total IgG avidity after the second injection.
B cell activation and maturation to antibody-secreting plasma cells take place in the germinal centers (GC) of lymphatic tissue. Messenger RNA vaccines efficiently promote the formation of germinal centers in vivo. In mice, the splenic GC showed a 10–20-fold increase of B cells upon nucleotide-modified mRNA/LNP immunization, significantly higher than after immunization with a recombinant protein or inactivated viral particles (Pardi et al. 2018). Enlarged GCs, pronounced increase in proliferating B cells and follicular helper T cells (TfH), were also observed in NHPs after immunization with nucleotide-modified mRNA/LNPs (Lindgren 2017). The frequency of antigen-specific TfH (4–8%) was significantly higher than observed after vaccination with protein (<1%) (Pardi et al. 2018). Since TfH cells are necessary for B cell affinity maturation and isotype switch, an increase in TfH cells should correlate positively with higher antibody titers (McHeyzer-Williams et al. 2009; Lindgren 2017). Finally, the first mature antibody-secreting plasma cells in the bone marrow were detected 2 weeks post prime immunization (Lindgren 2017). Low number of circulating memory B cells were also detectable. They significantly increased upon boost and remained stable for at least 25 weeks post immunization.
3.3.2 Cellular Immunity
In line with the ability to activate B cells, mRNA vaccines induce strong CD4+ and CD8+ T cell responses to various pathogens, e.g., rabies and influenza, including the establishment of effector memory CD8+ T cells (CD44+CCR7−CD62L−) (Fotin-Mleczek 2011; Lutz 2017; Petsch 2012). Several sequential immunizations boost T cell response further without a sign of T cell exhaustion or induction of regulatory T cells (Fotin-Mleczek 2011; Kowalczyk 2016). The original data used protamine-formulated mRNA, but new data with LNP-formulation confirmed efficient T cell induction by mRNA in mice and NHP, outperforming licensed vaccines (Lutz 2017; Petsch 2012).
The induction of a strong Th1 response is specifically preferred for vaccines against intracellular pathogens, since it not only leads to high antibody response but also induces cytotoxic T cells. T cells can eliminate infected cells and prevent further spread of a viral or different intracellular pathogen. Immunization with naked mRNA has been reported to induce Th2 responses, while LNP- or protamine-formulated mRNAs stimulate a Th1 response through the activation of TLRs and the signaling by MyD88 (Kranz et al. 2016; Fotin-Mleczek 2011; Scheel 2004; Carralot 2004; Pollard 2013). This is supported by the observation of higher IgG2a /IgG1 ratio in mice. After immunization with protamine-formulated mRNA, mainly IFNγ and IL-2 secreting CD4+ T cells were detected (Fotin-Mleczek 2011), which supports activation of CD8+ T cells. Pardi et al. reported induction of multifunctional CD4+ T cells expressing TNF, IFNγ, and IL-2 upon two i.d. immunizations of nucleotide-modified mRNA/LNP (Pardi et al. 2018).
mRNA vaccines induce cellular immunity more efficiently than standard licensed vaccines and stimulate multifunctional effector T cells of multiple subsets.
4 Conclusion
The first proof of concept for an mRNA vaccine was reported two decades ago (Martinon et al. 1993; Hoerr et al. 2000). Since mRNA vaccines against a multitude of pathogens have been developed and characterized in pre-clinical models. mRNA vaccines have several advantages over recombinant protein and whole viral particle vaccines. ssRNA itself has an adjuvant-effect, which abolishes the need of an additional adjuvant (Edwards 2017). The antigen expressed by the host cells facilitates correct folding and conformation and enables incorporation of transmembrane proteins to be presented on the cell surface. Viral antigens are expressed in situ similarly to expression during viral infection. Due to the lack of host gene integration risk, mRNA vaccines promise a better safety profile than DNA vectors. mRNA technology enables broad infectious disease application. It is independent of pathogen cultivation and inactivation and hence does not require specific biosafety environment. The basic manufacturing process for different vaccines is similar and does not need major adaptation to different pathogens (Schlake et al. 2012). The process can be streamlined and adjusted to global health threats. Recent advances in understanding the influence of untranslated mRNA sequences, formulations, and injection technologies suggest new and exciting developments in the field of mRNA vaccines in the next few years. New technologies, including machine learning and artificial intelligence, will provide new insights in mRNA vaccine designs for improved product candidates.
The proof of concept of mRNA vaccines was already demonstrated for several viral pathogens, e.g., influenza virus, rabies, CMV, ZIKA, HIV, tick-transmitted flaviviruses (Richner 2017; Pardi et al. 2017, 2018, 2019; Lutz 2017; Petsch 2012; Schnee 2016; John 2018; John et al. 2018).
Very high and long-lasting antibody titers are detected in both rodents and NHPs. The strong T cell response indicates the opportunity for therapeutic vaccination against chronic diseases such as life-long pathogenic infections or cancer. mRNA vaccines are immunogenic and protective in various animal models. Multiple clinical trials are ongoing, e.g., rabies virus (NCT03713086), CMV (NCT03382405), influenza (NCT03345043), as well as at least three mRNA vaccines for the ongoing SARS-CoV-2 pandemic outbreak (NCT04283461, NCT04449276, NCT04470427, NCT04405076, NCT04368728, NCT04380701). This highlights that especially in the case of a pandemic, mRNA vaccines are among the first vaccines manufactured for clinical use and subject to clinical testing. Already four months after the SARS-CoV-2 pandemic outbreak in December 2019, an mRNA vaccine platform was the first vaccine used in a clinical trial starting 16th March (NCT04283461). The first clinical batch (https://www.modernatx.com/modernas-work-potential-vaccine-against-covid-19) was available 25 days after sequence selection. Efficacy was demonstrated for two mRNA vaccines within eleven months for two independent mRNA vaccines for SARS-CoV-2 in large phase III efficacy studies (NCT04368728, NCT04470427), and conditional marketing applications have been submitted by two companies to competent authorities. This unprecedented speed of development underscores that mRNA vaccines will have their share in preventing disease and addressing unmet medical need, including the ongoing Covid-19 pandemic and are proofing their value for future pandemic preparedness.
References
Ahmed F, Benedito VA, Zhao PX (2011) Mining functional elements in messenger RNAs: overview, challenges, and perspectives. Front Plant Sci 2
Alberer M et al (2017) Safety and immunogenicity of a mRNA rabies vaccine in healthy adults: an open-label, non-randomised, prospective, first-in-human phase 1 clinical trial. Lancet
Allen IC et al (2009) The NLRP3 inflammasome mediates in vivo innate immunity to influenza A virus through recognition of viral RNA. Immunity 30(4):556–565
Anderson BR et al (2010) Incorporation of pseudouridine into mRNA enhances translation by diminishing PKR activation. Nucleic Acids Res 38(17):5884–5892
Anderson BR, Muramatsu H, Jha BK, Silverman RH, Weissman D, Karikó K (2011) Nucleoside modifications in RNA limit activation of 2ʹ-5ʹ-oligoadenylate synthetase and increase resistance to cleavage by RNase L. Nucleic Acids Res 39(21):9329–9338
Andries O et al (2013) Innate immune response and programmed cell death following carrier-mediated delivery of unmodified mRNA to respiratory cells. J Control Release 167(2):157–166
Andries O, Mc Cafferty S, De Smedt SC, Weiss R, Sanders NN, Kitada T (2015) N1-methylpseudouridine-incorporated mRNA outperforms pseudouridine-incorporated mRNA by providing enhanced protein expression and reduced immunogenicity in mammalian cell lines and mice. J Control Release 217:337–344
Bahl K et al (2017) Preclinical and clinical demonstration of immunogenicity by mRNA vaccines against H10N8 and H7N9 influenza viruses. Mol Ther 25(6):1316–1327
Baiersdörfer M et al (2019) A facile method for the removal of dsRNA contaminant from in vitro-transcribed mRNA. Mol Ther Nucleic Acids 15:26–35
Banerjee AK (1980) 5’-terminal cap structure in eucaryotic messenger ribonucleic acids. Microbiol Rev 44(2):175–205
Beddows S et al (2006) Construction and characterization of soluble, cleaved, and stabilized trimeric env proteins Based on HIV type 1 env subtype A. AIDS Res Hum Retroviruses 22(6):569–579
Ben-Asouli Y, Banai Y, Pel-Or Y, Shir A, Kaempfer R (2002) Human interferon-γ mRNA autoregulates its translation through a Pseudoknot that activates the interferon-inducible protein kinase PKR. Cell 108(2):221–232
Berkovits BD, Mayr C (2015) Alternative 3ʹ UTRs act as scaffolds to regulate membrane protein localization. Nature 522(7556):363–367
Bernstein P, Peltz SW, Ross J (1989) The poly(A)-poly(A)-binding protein complex is a major determinant of mRNA stability in vitro. Mol. Cell. Biol. 9(2):659–670
Beverley PCL (2002) Immunology of vaccination. Br Med Bull 62(1):15–28
Beverly M, Hagen C, Slack O (2018) Poly A tail length analysis of in vitro transcribed mRNA by LC-MS. Anal Bioanal Chem 410(6):1667–1677
Borg FA, Isenberg DA (2007) Syndromes and complications of interferon therapy. Curr Opin Rheumatol 19(1):61–66
Bourhy H et al (2007) “Annex 2 Recommendations for inactivated rabies vaccine for human use produced in cell substrates and embryonated eggs. World Heal Organ Tech Rep Ser 941:83–132
Brisse M, Ly H (2019) Comparative structure and function analysis of the RIG-I-like receptors: RIG-I and MDA5. Front Immunol 10
Broos K et al (2016) Particle-mediated Intravenous Delivery of Antigen mRNA Results in Strong Antigen-specific T-cell Responses Despite the Induction of Type I Interferon. Mol. Ther. - Nucleic Acids 5:e326
Burns CC, Diop OM, Sutter RW, Kew OM (2014) Vaccine-derived polioviruses. J Infect Dis 210(suppl 1):S283–S293
Burton DR, Hangartner L (2016) Broadly neutralizing antibodies to HIV and their role in vaccine design. Annu Rev Immunol 34:635–659
Carr MW, Roth SJ, Luther E, Rose SS, Springer TA (1994) Monocyte chemoattractant protein 1 acts as a T-lymphocyte chemoattractant. Proc Natl Acad Sci USA 91(9):3652
Carralot JP et al (2004) Polarization of immunity induced by direct injection of naked sequence-stabilized mRNA vaccines. Cell Mol Life Sci 61(18):2418–2424
Chumakov K, Elzinga N, Wood DJ (2002) Annex 2 Recommendations for the production and control of poliomyelitis vaccine ( inactivated) 1. World Heal Organ Tech Rep Ser (910)
Clarke TF IV, Clark PL (2010) Increased incidence of rare codon clusters at 5ʹ and 3ʹ gene termini: implications for function. BMC Genomics 11:118
Crouse J, Kalinke U, Oxenius A (2015) Regulation of antiviral T cell responses by type I interferons. Nat Rev Immunol 15(4):231–242
De Beuckelaer A et al (2016) Type I interferons interfere with the capacity of mRNA lipoplex vaccines to elicit cytolytic T cell responses. Mol Ther 24(11):2012–2020
De Beuckelaer A, Grooten J, De Koker S (2017) Type I interferons modulate CD8+ T cell immunity to mRNA vaccines. Trends Mol Med 23(3):216–226
Devarkar SC et al (2016) Structural basis for m7G recognition and 2′-O-methyl discrimination in capped RNAs by the innate immune receptor RIG-I. Proc Natl Acad Sci USA 113(3):596
Devoldere J, Dewitte H, De Smedt SC, Remaut K (2016) Evading innate immunity in nonviral mRNA delivery: don’t shoot the messenger. Drug Discov Today 21(1):11–25
Ding Y, Tang Y, Kwok CK, Zhang Y, Bevilacqua PC, Assmann SM (2014) In vivo genome-wide profiling of RNA secondary structure reveals novel regulatory features. Nature 505(7485):696–700
Durbin AF, Wang C, Marcotrigiano J, Gehrke L (2016) RNAs containing modified nucleotides fail to trigger RIG-I conformational changes for innate immune signaling. MBio 7(5)
Edwards et al DK (2017) Adjuvant effects of a sequence-engineered mRNA vaccine: translational profiling demonstrates similar human and murine innate response. J Transl Med 15(1):1
Elango N, Elango S, Shivshankar P, Katz MS (2005) Optimized transfection of mRNA transcribed from a d(A/T)100 tail-containing vector. Biochem Biophys Res Commun 330(3):958–966
Elfakess R, Sinvani H, Haimov O, Svitkin Y, Sonenberg N, Dikstein R (2011) Unique translation initiation of mRNAs-containing TISU element. Nucleic Acids Res 39(17):7598–7609
Fensterl V, Sen GC (2015) Interferon-induced Ifit proteins: their role in viral pathogenesis. J Virol 89(5):2462
Ford LP, Bagga PS, Wilusz J (1997) The poly(A) tail inhibits the assembly of a 3ʹ-to-5ʹ exonuclease in an in vitro RNA stability system. Mol Cell Biol 17(1):398–406
Fotin-Mleczek M et al (2011) Messenger RNA-based vaccines with dual activity induce balanced TLR-7 dependent adaptive immune responses and provide antitumor activity. J Immunother 34(1):1–15
Galloway A, Cowling VH (2019) mRNA cap regulation in mammalian cell function and fate. Biochim Biophys Acta Gene Regul Mech 1862(3):270
Geall AJ et al (2012) Nonviral delivery of self-amplifying RNA vaccines. Proc Natl Acad Sci 109(36):14604–14609
Georg P, Sander LE (2019) Innate sensors that regulate vaccine responses. Curr Opin Immunol 59:31–41
Grier AE et al (2016) pEVL: a linear plasmid for generating mRNA IVT templates with extended encoded poly(A) sequences. Mol Ther Nucleic Acids 5(4):e306
Groom JR, Luster AD (2011) CXCR3 ligands: redundant, collaborative and antagonistic functions. Immunol Cell Biol 89(2)
Grudzien-Nogalska E, Jemielity J, Kowalska J, Darzynkiewicz E, Rhoads RE (2007) Phosphorothioate cap analogs stabilize mRNA and increase translational efficiency in mammalian cells. RNA 13(10):1745–1755
Gruys E, Toussaint MJM, Niewold TA, Koopmans SJ (2005) Acute phase reaction and acute phase proteins. J Zhejiang Univ Sci B 6(11):1045–1056
Gustafsson C, Govindarajan S, Minshull J (2004) Codon bias and heterologous protein expression. Trends Biotechnol 22(7):346–353
Haralambieva IH, Kennedy RB, Ovsyannikova IG, Schaid DJ, Poland GA (2019) Current perspectives in assessing humoral immunity after measles vaccination. Expert Rev Vaccines 18(1):75
Hesseling A et al (2009) Disseminated bacille Calmette-Guérin disease in HIV-infected South African infants. Bull World Health Organ 87(7):505
Hickling J, Jones K, Friede M, Zehrung D, Chen D, Kristensen D (2011) Intradermal delivery of vaccines: potential benefits and current challenges. Bull World Health Organ 89(3):221
Hoerr I, Obst R, Rammensee HG, Jung G (2000) In vivo application of RNA leads to induction of specific cytotoxic T lymphocytes and antibodies. Eur J Immunol 30(1):1–7
Holtkamp S et al (2006) Modification of antigen-encoding RNA increases stability, translational efficacy and T-cell stimularoy capacity of dendritic cells. Blood 108(13)
Iavarone C, O’hagan DT, Yu D, Delahaye NF, Ulmer JB (2017) Mechanism of action of mRNA-based vaccines. Expert Rev Vaccines 16(9):871–881
Innis BL et al (1994) Protection against hepatitis A by an inactivated vaccine. JAMA J Am Med Assoc 271(17):1328
Jackson LA et al (2020) An mRNA vaccine against SARS-CoV-2—preliminary report. N Engl J Med
Jan CH, Friedman RC, Ruby JG, Bartel DP (2011) Formation, regulation and evolution of Caenorhabditis elegans 3′ UTRs. Nature 469(7328):97–101
Jemielity J et al (2003) Novel ‘anti-reverse’ cap analogs with superior translational properties. RNA 9(9):1108–1122
Jia J, Yao P, Arif A, Fox PL (2013) Regulation and dysregulation of 3ʹ UTR-mediated translational control. Curr Opin Genet Dev 23(1):29–34
John S et al (2018) Multi-antigenic human cytomegalovirus mRNA vaccines that elicit potent humoral and cell-mediated immunity. Vaccine 36(12):1689–1699
Juskewitch JE, Tapia CJ, Windebank AJ (2010) Lessons from the Salk polio vaccine: methods for and risks of rapid translation. Clin Transl Sci 3(4):182–185
Karikó K, Buckstein M, Ni H, Weissman D (2005) Suppression of RNA recognition by toll-like receptors: the impact of nucleoside modification and the evolutionary origin of RNA. Immunity 23(2):165–175
Karikó K et al (2008) Incorporation of pseudouridine into mRNA yields superior nonimmunogenic vector with increased translational capacity and biological stability. Mol Ther 16(11):1833–1840
Karikó K, Muramatsu H, Ludwig J, Weissman D (2011) Generating the optimal mRNA for therapy: HPLC purification eliminates immune activation and improves translation of nucleoside-modified, protein-encoding mRNA. Nucleic Acids Res 39(21):e142
Karikó K, Muramatsu H, Keller JM, Weissman D (2012) Increased erythropoiesis in mice injected with submicrogram quantities of pseudouridine-containing mRNA encoding erythropoietin. Mol Ther 20(5):948–953
Kato H et al (2008) Length-dependent recognition of double-stranded ribonucleic acids by retinoic acid-inducible gene-I and melanoma differentiation-associated gene 5. J Exp Med 205(7):1601–1610
Kauffman KJ et al (2016) Efficacy and immunogenicity of unmodified and pseudouridine-modified mRNA delivered systemically with lipid nanoparticles in vivo. Biomaterials 109:78–87
Kaygun H, Marzluff WF (2005) Translation termination is involved in histone mRNA degradation when DNA replication is inhibited. Mol Cell Biol 25(16):6879–6888
Kierzek E, Kierzek R (2003) The thermodynamic stability of RNA duplexes and hairpins containing N6-alkyladenosines and 2-methylthio-N6-alkyladenosines. Nucleic Acids Res 31(15):4472–4480
Kocmik I et al (2018) Modified ARCA analgos providing enhanced properties of capped mRNAs. Cell Cycle 17(13):71–75
Koh WS, Porter JR, Batchelor E (2019) Tuning of mRNA stability through altering 3’-UTR sequences generates distinct output expression in a synthetic circuit driven by p53 oscillations. Sci Rep 9(1):5976
Kore AR, Charles I (2010) Synthesis and evaluation of 2′–O–allyl substituted dinucleotide cap analog for mRNA translation. Bioorganic Med Chem 18(22):8061–8065
Kore AR, Shanmugasundaram M, Charles I, Vlassov AV, Barta TJ (2009) Locked nucleic acid (LNA)-modified dinucleotide mRNA cap analogue: synthesis, enzymatic incorporation, and utilization. J Am Chem Soc 131(18):6364–6365
Kormann MSD et al (2011) Expression of therapeutic proteins after delivery of chemically modified mRNA in mice. Nat Biotechnol 29(2):110–112
Kowalczyk A et al (2016) Self-adjuvanted mRNA vaccines induce local innate immune responses that lead to a potent and boostable adaptive immunity. Vaccine 34(33):3882–3893
Kozak M (2005) Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene 361(1–2):13–37
Kozak M (1991a) A short leader sequence impairs the fidelity of initiation by eukaryotic ribosomes. Gene Expr 1(2):111–115
Kozak M (1991b) Effects of long 5ʹ leader sequences on initiation by eukaryotic ribosomes in vitro. Gene Expr 1(2):117–125
Kranz LM et al (2016) Systemic RNA delivery to dendritic cells exploits antiviral defence for cancer immunotherapy. Nature 534
Kuhn AN et al (2010) Phosphorothioate cap analogs increase stability and translational efficiency of RNA vaccines in immature dendritic cells and induce superior immune responses in vivo. Gene Ther 17(8):961–971
Kuhn AN, Diken M, Kreiter S, Vallazza B, Türeci Ö, Sahin U (2011) Determinants of intracellular RNA pharmacokinetics: Implications for RNA-based immunotherapeutics. RNA Biol 8(1):35–43
Lässig C, Hopfner K-P (2017) Discrimination of cytosolic self and non-self RNA by RIG-I-like receptors. J Biol Chem 292(22):9000–9009
Lawrence JB, Singer RH (1986) Intracellular localization of messenger RNAs for cytoskeletal proteins. Cell 45(3):407–415
Le Bon A et al (2014) Direct stimulation of T cells by type I IFN enhances the CD8+ T cell response during cross-priming. J Immunol 176(8):4682–4689
Lee J, Arun Kumar S, Jhan YY, Bishop CJ (2018) Engineering DNA vaccines against infectious diseases. Acta Biomater 80:31–47
Lemckert AAC et al (2005) Immunogenicity of heterologous prime-boost regimens involving recombinant adenovirus serotype 11 (Ad11) and Ad35 vaccine vectors in the presence of anti-Ad5 immunity. J Virol 79(15):9694–9701
Leppek K, Das R, Barna M (2018) Functional 5′ UTR mRNA structures in eukaryotic translation regulation and how to find them. Nat Rev Mol Cell Biol 19(3):158–174
Liang F et al (2017) Efficient targeting and activation of antigen-presenting cells in vivo after modified mRNA vaccine administration in rhesus macaques. Mol Ther 25(12):2635–2647
Lindgren G et al (2017) Induction of robust B cell responses after influenza mRNA vaccination is accompanied by circulating hemagglutinin-specific ICOS+ PD-1+ CXCR3+ T follicular helper cells. Front Immunol 8:1539
Loomis KH et al (2018) In vitro transcribed mRNA vaccines with programmable stimulation of innate immunity. Bioconjug Chem 29(9):3072–3083
Lutz J et al (2017) Unmodified mRNA in LNPs constitutes a competitive technology for prophylactic vaccines. NPJ Vaccines 2(1):1–9
Mannironi C, Bonner WM, Hatch CL (1989) H2A.X. a histone isoprotein with a conserved C-terminal sequence, is encoded by a novel mRNA with both DNA replication type and polyA 3ʹ processing signals. Nucleic Acids Res 17(22):9113
Mariner JC et al (2012) Rinderpest eradication: appropriate technology and social innovations. Science (80–) 337 (6100):1309–1312
Martinon F et al (1993) Induction of virus-specific cytotoxic T lymphocytesin vivo by liposome-entrapped mRNA. Eur J Immunol 23(7):1719–1722
Marzluff WF (1992) Histone 3ʹ ends: essential and regulatory functions. Gene Expr 2(2)
Mauger DM et al (2019) mRNA structure regulates protein expression through changes in functional half-life. Proc Natl Acad Sci USA 116(48):24075–24083
Mayr C (2008) Regualtion by 3ʹ-untranslated regions. Postgrad Med J 67(791):862–862
McHeyzer-Williams LJ, Pelletier N, Mark L, Fazilleau N, McHeyzer-Williams MG (2009) Follicular helper T cells as cognate regulators of B cell immunity. Curr Opin Immunol 21(3):266–273
McNab F, Mayer-Barber K, Sher A, Wack A, O’Garra A (2015) Type I interferons in infectious disease. Nat Rev Immunol 15(2):87–103
Meis R, Meis JE, Biotechnologies E (2006) Achieve 100 % capping efficiency with the NEW ScriptCapTM m 7 G capping system improve the translation efficiency of any 5ʹ-capped mRNA with the NEW ScriptCapTM 2ʹ–O–Methyltransferase. System 13(4):5–6
Meis JE, Meis R, Biotechnologies E (2016) The new mScript TM mRNA production system—efficient mRNA transcription, capping and tailing for the highest yields of active protein. Epic Biotechnol Forum 14(1):4–5
Mockey M, Gonçalves C, Dupuy FP, Lemoine FM, Pichon C, Midoux P (2006) mRNA transfection of dendritic cells: synergistic effect of ARCA mRNA capping with Poly(A) chains in cis and in trans for a high protein expression level. Biochem Biophys Res Commun 340(4):1062–1068
Mogensen TH (2009) Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev 22(2):240
Mu X, Greenwald E, Ahmad S, Hur S (2018) An origin of the immunogenicity of in vitro transcribed RNA. Nucleic Acids Res 46(10):5239–5249
Muckenthaler MU, Rivella S, Hentze MW, Galy B (2017) A red carpet for iron metabolism. Cell 168(3):344
Mulligan MJ et al (2020) Phase 1/2 study to describe the safety and immunogenicity of a COVID-19 RNA vaccine candidate (BNT162b1) in adults 18 to 55 years of age: interim report. medRxiv https://doi.org/10.1101/2020.06.30.20142570
Ndhlovu ZM et al (2015) The breadth of expandable memory CD8+ T cells inversely correlates with residual viral loads in HIV elite controllers. J Virol 89(21):10735–10747
Nigg AJ, Walker PL (2009) Overview, prevention, and treatment of rabies. Pharmacotherapy 29(10):1182–1195
Novoa EM, Ribas de Pouplana L (2012) Speeding with control: codon usage, tRNAs, and ribosomes. Trends Genet 28(11):574–581
Orlandini von Niessen AG et al (2019) Improving mRNA-based therapeutic gene delivery by expression-augmenting 3ʹ UTRs identified by cellular library screening. Mol Ther 27(4):824–836
Ozawa S et al (2017) Estimated economic impact of vaccinations in 73 low- and middle-income countries, 2001–2020. Bull World Health Organ 95(9):629
Padilla-Quirarte HO, Lopez-Guerrero DV, Gutierrez-Xicotencatl L, Esquivel-Guadarrama F (2019) Protective antibodies against influenza proteins. Front Immunol 10
Pardi N et al (2017a) Administration of nucleoside-modified mRNA encoding broadly neutralizing antibody protects humanized mice from HIV-1 challenge. Nat Commun 8:14630
Pardi N et al (2017b) Zika virus protection by a single low-dose nucleoside-modified mRNA vaccination. Nature 543(7644):248–251
Pardi N et al (2018) Nucleoside-modified mRNA immunization elicits influenza virus hemagglutinin stalk-specific antibodies. Nat Commun 9(1):3361
Pardi N et al (2018) Nucleoside-modified mRNA vaccines induce potent T follicular helper and germinal center B cell responses. J Exp Med 215(6):1571–1588
Pardi N et al (2019) Characterization of HIV-1 nucleoside-modified mRNA vaccines in rabbits and rhesus macaques. Mol Ther Nucleic Acids 15:36–47
Pasquinelli A, Dahlberg J, Elsebet L (1995) Reverse 5’ caps in RNAs made in vitro by phage RNA polymerases. RNA 1(957):110–117
Patinote C et al (2020) Agonist and antagonist ligands of toll-like receptors 7 and 8: ingenious tools for therapeutic purposes. Eur J Med Chem 193:112238
Petrovsky N (2015) Comparative safety of vaccine adjuvants: a summary of current evidence and future needs. Drug Saf 38(11):1059–1074
Petsch B et al (2012) Protective efficacy of in vitro synthesized, specific mRNA vaccines against influenza A virus infection. Nat Biotechnol 30(12):1210–1216
Plotkin S (2014) History of vaccination. Proc Natl Acad Sci USA 111(34):12283
Pollard C et al (2013) Type I IFN counteracts the induction of antigen-specific immune responses by lipid-based delivery of mRNA vaccines. Mol Ther 21(1):251–259
Rabani M, Pieper L, Chew GL, Schier AF (2017) A massively parallel reporter assay of 3′ UTR sequences identifies in vivo rules for mRNA degradation. Mol Cell 68(6):1083-1094.e5
Rajan JV, Warren SE, Miao EA, Aderem A (2010) Activation of the NLRP3 inflammasome by intracellular poly I:C. FEBS Lett 584(22):4627–4632
Rauch S, Jasny E, Schmidt KE, Petsch B (2018) New vaccine technologies to combat outbreak situations. Front Immunol 9:1963
Richner JM et al (2017) Modified mRNA vaccines protect against Zika virus infection. Cell 168(6):1114-1125.e10
Rönnblom LE (1991) Autoimmunity after alpha-interferon therapy for malignant carcinoid tumors. Ann Intern Med 115(3):178
Rossey I, McLellan JS, Saelens X, Schepens B (2018) Clinical potential of prefusion RSV F-specific antibodies. Trends Microbiol 26(3):209–219
Rydzik AM et al (2017) mRNA cap analogues substituted in the tetraphosphate chain with CX2: identification of O-to-CCl2 as the first bridging modification that confers resistance to decapping without impairing translation. Nucleic Acids Res 45(15):8661–8675
Sadler AJ, Williams BRG (2008) Interferon-inducible antiviral effectors. Nat Rev Immunol 8(7):559–568
Sample PJ et al (2019) Human 5′ UTR design and variant effect prediction from a massively parallel translation assay. Nat Biotechnol 37(7):803
Sanders RW, Moore JP (2017) Native-like env trimers as a platform for HIV-1 vaccine design. Immunol Rev 275(1):161
Scheel B et al (2004) Immunostimulating capacities of stabilized RNA molecules. Eur J Immunol 34(2):537–547
Schlake T, Thess A, Fotin-mleczek M, Kallen K (2012) Developing mRNA-vaccine technologies. RNA Biol 9(November):1–12
Schlake T, Thess A, Thran M, Jordan I (2019) mRNA as novel technology for passive immunotherapy. Cell Mol Life Sci 76(2):301–328
Schmidt A et al (2009) 5′-triphosphate RNA requires base-paired structures to activate antiviral signaling via RIG-I. Proc Natl Acad Sci USA 106(29):12067
Schnee M et al (2016) An mRNA vaccine encoding rabies virus glycoprotein induces protection against lethal infection in mice and correlates of protection in adult and newborn pigs. PLoS Negl Trop Dis 10(6):e0004746
Schnierle BS, Gershon PD, Moss B (1992) Cap-specific mRNA (nucleoside-O2ʹ-)-methyltransferase and poly(A) polymerase stimulatory activities of vaccinia virus are mediated by a single protein. Proc Natl Acad Sci USA 89(7):2897–2901
Shanmugasundaram M, Charles I, Kore AR (2016) Design, synthesis and biological evaluation of dinucleotide mRNA cap analog containing propargyl moiety. Bioorganic Med Chem 24(6):1204–1208
Skowronski DM et al (2014) Low 2012–13 influenza vaccine effectiveness associated with mutation in the egg-adapted H3N2 vaccine strain not antigenic drift in circulating viruses. PLoS ONE 9(3):e92153
Spenkuch F, Motorin Y, Helm M (2014) Pseudouridine: still mysterious, but never a fake (uridine)! RNA Biol 11(12):1540
Stepinski J, Waddell C, Stolarski R, Darzynkiewicz E, Rhoads RE (2001) Synthesis and properties of mRNAs containing the novel ‘anti-reverse’ cap analogs 7-methyl(3’-O-methyl)GpppG and 7-methyl (3’-deoxy)GpppG. RNA 7(10):1486–1495
Stewart M (2019) Polyadenylation and nuclear export of mRNAs. J Biol Chem 294(9):2977–2987
Stitz L et al (2017) A thermostable messenger RNA based vaccine against rabies. PLoS Negl Trop Dis 11(12):1–10
Svitkin YV, Cheng YM, Chakraborty T, Presnyak V, John M, Sonenberg N (2017) N1-methyl-pseudouridine in mRNA enhances translation through eIF2α-dependent and independent mechanisms by increasing ribosome density. Nucleic Acids Res 45(10):6023–6036
Swiecki M, Wang Y, Vermi W, Gilfillan S, Schreiber RD, Colonna M (2011) Type I interferon negatively controls plasmacytoid dendritic cell numbers in vivo. J Exp Med 208(12):2367–2374
Tannenbaum CS, Tubbs R, Armstrong D, Finke JH, Bukowski RM, Hamilton TA (1998) The CXC Chemokines IP-10 and Mig are necessary for IL-12-mediated regression of the mouse RENCA tumor. J Immunol 153(10):4625–4635
Thess A et al (2015) Sequence-engineered mRNA without chemical nucleoside modifications enables an effective protein therapy in large animals. Mol Ther 23(9):1456–1464
Trepotec Z, Aneja MK, Geiger J, Hasenpusch G, Plank C, Rudolph C (2018) Maximizing the translational yield of mRNA therapeutics by minimizing 5′-UTRs. Tissue Eng Part A 25(1–2):69–79
Trepotec Z, Geiger J, Plank C, Aneja MK, Rudolph C (2019) Segmented poly(A) tails significantly reduce recombination of plasmid DNA without affecting mRNA translation efficiency or half-life. RNA 25(4):507–518
Tuller T, Zur H (2015) Multiple roles of the coding sequence 5′ end in gene expression regulation. Nucleic Acids Res 43(1):13
Tusup M, French LE, Guenova E, Kundig T, Pascolo S (2018) Optimizing the functionality of in vitro-transcribed mRNA. Biomed J Sci Tech Res 7(2):5845–5850
Uchida S, Kataoka K, Itaka K (2015) Screening of mRNA chemical modification to maximize protein expression with reduced immunogenicity. Pharmaceutics 7(3):137–151
Vaidyanathan S et al (2018) Uridine depletion and chemical modification increase Cas9 mRNA activity and reduce immunogenicity without HPLC purification. Mol Ther Nucleic Acids 12:530
Vanblargan LA et al (2018) An mRNA vaccine protects mice against multiple article an mRNA vaccine protects mice against multiple tick-transmitted flavivirus infections, pp. 3382–3392
Venkatesan S, Gershowitz A, Moss B (1980) Modification of the 5ʹ end of mRNA. J Biol Chem 255(3):903–908
Vivinus S et al (2001) An element within the 5′ untranslated region of human Hsp70 mRNA which acts as a general enhancer of mRNA translation. Eur J Biochem 268(7):1908–1917
Wang Z, Day N, Trifillis P, Kiledjian M (1999) An mRNA stability complex functions with poly(A)-binding protein to stabilize mRNA in vitro. Mol Cell Biol 19(7):4552–4560
Wills RJ, Dennis S, Spiegel HE, Gibson DM, Nadler PI (1984) Interferon kinetics and adverse reactions after intravenous, intramuscular, and subcutaneous injection. Clin Pharmacol Ther 35(5):722–727
Wolff JA et al (1990) Direct gene transfer into mouse muscle in vivo. Science 247(4949 Pt 1):1465–1468
Wolpe SD, Cerami A (1989) Macrophage inflammatory proteins 1 and 2: members of a novel superfamily of cytokines. FASEB J 3(14)
World Health Organization (1980) World Health Assembly, 33. Declaration of global eradication of smallpox
Zhang L, Wang W, Wang S (2015) Effect of vaccine administration modality on immunogenicity and efficacy. Expert Rev Vaccines 14(11):1509–1523
Zost SJ et al (2017) Contemporary H3N2 influenza viruses have a glycosylation site that alters binding of antibodies elicited by egg-adapted vaccine strains. Proc Natl Acad Sci USA 114(47):12578–12583
Acknowledgements
We thank Igor Splawski and Janine Mühe for critically reading the manuscript.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2020 Springer Nature Switzerland AG
About this chapter
Cite this chapter
Gergen, J., Petsch, B. (2020). mRNA-Based Vaccines and Mode of Action. In: Yu, D., Petsch, B. (eds) mRNA Vaccines. Current Topics in Microbiology and Immunology, vol 440. Springer, Cham. https://doi.org/10.1007/82_2020_230
Download citation
DOI: https://doi.org/10.1007/82_2020_230
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-18069-9
Online ISBN: 978-3-031-18070-5
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)