Abstract
Although the first natural products (NP) from Photorhabdus and Xenorhabdus bacteria have been known now for almost 30 years, a huge variety of new compounds have been identified in the last 5–10 years, mainly due to the application of modern mass spectrometry. Additionally, application of molecular methods that allow the activation of NP production in several different strains as well as efficient heterologous expression methods have led to the production and validation of many new compounds. In this chapter we discuss the benefit of using Photorhabdus as a model system for microbial chemical ecology. We also examine non-ribosomal peptide synthetases as the most important pathway for NP production. Finally, we discuss the origin and function of all currently known NPs and the development of the molecular and chemical tools used to identify these NPs faster.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Natural Product
- Peptide Bond Formation
- Biosynthesis Gene Cluster
- Promoter Exchange
- Entomopathogenic Bacterium
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Low molecular weight natural products (NPs) from plants and other biological sources have been used by humans since ancient times. Today’s modern life without such microbial natural products would be nearly impossible. For example, 30–60 % of all drugs currently in clinical use as antibiotics (e.g. vancomycin, daptomycin, penicillin, erythromycin), anti-cancer agents (e.g. doxycycline) or immune suppressant compounds (e.g. cyclosporine, rapamycin) are natural products or are directly derived from them (Newman and Cragg 2012). Despite our success in identifying these compounds and their subsequent medical utility it is still not clear why these natural products are actually produced by these microorganisms and therefore what role(s) they play in their biology. Thus, whilst it is easy to infer a role for antibiotically active NPs in growth competition and (or) defence in the natural environment. For other natural products, such as those used as cholesterol lowering agents, their biological role(s) in nature are far less clear. In this respect it is interesting to note that even ‘antibiotics’ induce changes in the mRNA and protein levels of other bacteria when used in sub-inhibitory concentrations (Davies and Ryan 2012; Davies et al. 2006). Therefore compounds used by us as antibiotics might in fact assume a ‘signalling’ function at the low concentrations found in the natural environments. Only the subsequent ‘misuse’ of these signalling compounds in non-natural high doses then might result in a clinically relevant antibiotic activity.
Originally, NPs from microorganisms and plants were designated as ‘secondary’ metabolites not essential for growth in contrast to the ‘primary’ metabolites considered essential for growth. The limits of this primary–secondary classification were always the specific laboratory conditions used for such analysis. For example, typically a pure culture of a single bacterium was studied without any competitor in the laboratory. However, subsequent studies of microbial ecology have revealed that typical secondary metabolites may indeed have an ‘essential’ role but only under specific conditions of growth and or competition (e.g. iron siderophores or quorum sensing molecules). Thus, it might be more reasonable to call such compounds ‘specialized’ metabolites for specific conditions we often either do not recognize or fail to classify properly (Sharon et al. 2014).
From the recent genome sequencing projects of various microorganisms it has become obvious that almost bacteria, including anaerobes, that have previously been thought to not encode any biosynthesis gene cluster (BGC) for NP production do indeed carry such BGCs (Bode and Müller 2005). In fact between 6 and 8 % of the genome can be dedicated to NP biosynthesis with up to 25 BGCs found in some Streptomyces strains (Ikeda et al. 2003; Omura et al. 2001). Despite the extent of the genome encoding NPs, most of them are not expressed under the conditions analysed and are therefore often called ‘silent’. This point will be addressed in detail in the chapter by David Clarke also in this volume. Because of the large amount of energy and resources used to maintain NP production, with several microbes producing multiple compounds in parallel (Bode and Müller 2005), there is increasing evidence that NP production is regulated ‘heterogeneously’, as also found for the production of some quorum sensing compounds. In such a scenario, typical for a ‘division of labour’ (Ackermann 2015), only a fraction of cells actually produce a specific NP thereby reducing the workload for each single cell but also allowing the whole population to produce all NPs required at a specific time. Although, such research has just started it is evident from the sheer physical size of some NP BGCs that such a strategy would be extremely beneficial.
Probably the biggest current challenge in natural product research is the identification of the biological function of the specific NP produced. Despite recent rapid progress in microbial ecology, such as the development of single colony growth chambers and direct microbial detection (Ling et al. 2015), we are still limited by the complexity of natural communities and our ability to culture only a tiny fraction of this complex mixture of micro-organisms, in combination with a limited number of molecular tools to examine those organisms which we can indeed culture. Therefore, the real chemical ecology of NPs from typical soil microbes, like Streptomyces, is still a very complex black box leading people to turn to more simple systems to address basic questions about the biological role of NPs. In this respect, micro-organisms living in symbiosis might carry an advantage for such studies when all the interacting partners can be isolated and studied in the laboratory. Although such laboratory systems might not be fully ‘natural’ since a number of natural competitors might be absent, they do allow us to address at least one naturally relevant interaction in detail and possibly from both sides in cases where the symbiotic host is also accessible to analysis and manipulation. The ultimate goal would be to address such complex symbioses, like the interaction of humans with their microbiome, identifying the influence of individual NPs for the shaping of the microbiome itself and subsequent human developmental biology and even behaviour (Dorrestein et al. 2014). Since the human microbiome is still a major challenge, with a complexity similar to that seen in soil communities, people have started to look at lower eukaryote–microbe interactions as models (Cantley and Clardy 2015). In this respect, insects and nematodes are very promising model systems in which to tackle question of chemical ecology since they can both be cultivated easily in the laboratory and are also amenable to molecular methods.
In this chapter I will therefore focus on NPs from Photorhabdus bacteria which has been established as a model insect pathogen, with a few excursions to its sister taxon Xenorhabdus. I note that the main biosynthesis and regulation pathways for the production of Photorhabdus NPs will be introduced by David Clarke elsewhere in this volume.
2 Non-ribosomal Peptide Synthetases (NRPS) and Related Biosynthesis Pathways
Based on their biosynthetic pathway, peptide NPs can be divided into two classes (Grünewald and Marahiel 2006). Bacteriocines for instance are ribosomally synthesized and post-translationally modified peptides (RiPPS) with antibacterial activity often produced by bacteria to inhibit the growth of similar or closely related bacterial strains (Arnison et al. 2012). Further important subclass are non-ribosomally produced peptides, like the antibiotic daptomycin, the anti-cancer drug bleomycin and the immunosuppressant cyclosporin (Felnagle et al. 2008). The most striking difference to ribosomally synthesized peptides is their method of assembly. Non-ribosomal peptides (NRPs) are produced via large multi-enzyme complexes (mega-enzymes) using thiotemplate mechanisms like NRP synthetases (NRPS) and NRPS/PKS hybrids. They use peptide bond formation (NRPS) or a combination of peptide bond formation and Claisen-type condensation reactions (NRPS/polyketide synthase [PKS] hybrids) to build larger molecules from different building blocks, not limited to the 20 proteinogenic amino acids (Caboche et al. 2010). Further, these NRP exhibit unique structural elements, like D-, N- or C-methylated, glycosylated or phosphorylated amino acids, heterocycles, and/or N-terminal attached fatty acids.
A common feature of these molecules is their often constrained structure (Sieber and Marahiel 2005). For example, they exhibit macrocyclization (macrolactam or depsipeptide with one ester bond), heterocyclization or C–C or C–O cross-linking between different amino acids, which enhances the rigidity of their ring skeleton (Grünewald and Marahiel 2006). This rigidity in turn ensures bioactivity by a precise orientation required for interaction with a dedicated molecular target. Although peptides synthesized by NRPS are highly diverse in structure, most of them share a common mode of synthesis, denoted as multiple carrier thiotemplate mechanism. Further, NRPS harbour a strict ‘modular’ architecture. A ‘module’ is defined as the catalytic unit responsible for the incorporation of one specific building block (e.g. amino acid or AA) into the peptide chain that grows from the N- to the C-terminus and associated functional group modifications (Sieber and Marahiel 2005). Modules are composed of domains that catalyze the single reaction steps like activation, covalent binding, optional modification of the building blocks, and condensation with the amino acyl or peptidyl group on the neighbouring module (Mootz et al. 2002). At least three domains are necessary for the non-ribosomal production of peptides (see Fig. 1): An adenylation (A) domain, a peptidyl carrier protein (PCP) also denoted as thiolation (T) domain, and a condensation (C) domain (Grünewald and Marahiel 2006). These three domains are called ‘core’ domains.
The first (N-terminal) module (‘start’ module) of an NRPS often lacks the C domain, and the last (C-terminal) module (‘termination’ module) usually comprises a thioesterase (TE) domain (Sieber and Marahiel 2005). The TE domain is usually responsible for the release of linear (transfer to a water molecule), cyclic or branched cyclic peptides (amide or ester linkage). In addition to these ‘standard’ domains (C, A, T, TE), a cyclization (Cy) domain instead of a C domain and a terminal condensation (C term) domain in place of a TE domain may also be present. Further, modification domains like an epimerization (E) domain, N-methylation (MT) domain or oxidation (Ox) domain can also be part of such modules.
The reactions catalyzed by standard NRPS domains are now well understood (Marahiel 2016; Sieber and Marahiel 2005) and their essential enzymatic activities are illustrated in Fig. 2 (Mootz et al. 2002). These activities reside in the adenylation (A), thiolation (T) and condensation (C) domains. First, AAs are activated through the activity of an A domain (Fig. 2a). The energy derived from ATP hydrolysis is used to form an aminoacyl-adenylate intermediate. Then the AA is loaded on the thiol of the pantetheine cofactor of the T domain (Fig. 2b) (Gulick 2009). Consecutively activated AAs on T domains are then joined by a C domain that catalyzes peptide bond formation and transfers the upstream AA or peptide to the downstream substrate (Fig. 2c).
Essential enzymatic activities in non-ribosomal peptide synthases or NRPS. Active domains are shown in red. a Substrate recognition and activation by the A domain. b Covalent attachment of the activated aminoacyl adenylate onto the T domain bound 4’Ppan cofactor. c Peptide elongation. The C domain catalyzes an attack of the nucleophilic amine of the acceptor substrate onto the electrophilic thioester of the donor substrate. C domain’s acceptor site is indicated by ‘a’, the donor site by ‘d’
In most cases the number of NRPS modules corresponds directly to the number of AA residues incorporated into the associated peptide and the arrangement of the modules directly follows the peptides’ primary sequence because peptide synthesis proceeds collinear in an N- to C- terminal direction (Mootz et al. 2002). Such biosynthetic templates are called ‘linear’ or ‘type A’ NRPS (Fig. 3). Examples for linear NRPSs are the tyrocidine (Mootz and Marahiel 1997), surfactin (Cosmina et al. 1993) and GameXPeptide (Nollmann et al. 2015a) assembling NRPS.
In contrast to type A NRPSs, there are also ‘iterative’ or ‘type B’ and ‘nonlinear’ or ‘type C’ NRPSs (Mootz et al. 2002) (Fig. 5). Iterative NRPSs (e.g. enniatin, enterobactin) use their modules or domains more than once in the assembly of a single peptide. This strategy is employed to build up peptide chains that consist of shorter sequences that are repeated. Nonlinear NRPSs, such as vibriobactin, yersiniabactin and fungisporin, deviate in their domain organization from the standard (C-A-T) n module architecture (Mootz et al. 2002). They are characterized by at least one unusual arrangement of the core domains C, A and T, A deviation from the module arrangement (C-A-T) n of linear NRPSs often goes along with nonlinear peptide products that result from unusual internal cyclizations (e.g. bleomycin) or branch point syntheses (e.g. mycobactin). Consequently, the peptide sequence does not represent the linear module and domain arrangement of the biosynthetic template itself.
Two further cases must be considered regarding the role of inter-domain recognition in biosynthetic templates: (I) when domains are next to each other on the same polypeptide chain (domains acting in cis) or (II) when they belong to two distinct proteins (domains acting in trans), as NRPSs can be composed of several polypeptide chains (Lautru and Challis 2004). In the latter context, correct protein–protein recognition between NRPSs acting in trans is crucial to prevent unspecific interactions with non-partner enzymes and to guarantee an effective biosynthesis of the desired product, e.g. the tyrocidine biosynthetic systems consists of three distinct NRPSs (TycA, TycB and TycC) (Hahn and Stachelhaus 2004, 2006). The intermolecular communication within multienzyme NRPS complexes relies on the coordinated interplay of donor and acceptor communication-mediating (COM) domains (Chiocchini et al. 2006). In various studies, it has been shown that matching pairs of donor and acceptor domains promote the correct positioning of enzymes within multi-enzyme complexes and the selective translocation of intermediates between adjacent synthetases (Hahn and Stachelhaus 2004, 2006). COM domains are composed of 15–30 AA located at the C- and N-terminal end of the corresponding polypetide chains and possess α-helical structures that provide the interfaces for the selective differentiation between partner and non-partner NRPSs. In the specific case of multifunctional but single-protein NRPSs, when domains on the same polypeptide chain are interconnected through short stretches of amino acid residues, domain communication is acting in cis via defined catalytically inactive linker regions (~15 AA) (Lautru and Challis 2004). Studies on many systems have therefore provided compelling evidence that linkers are more than ‘simple’ covalent connectors (Gokhale and Khosla 2000).
3 Overview on Biosynthesis Gene Clusters and Natural Products in Different Photorhabdus Strains
A detailed analysis of the genome sequence of seven Photorhabdus strains (P. luminescens TTO1, P. luminescens PB45.5, P. asymbiotica ATCC 43949, P. asymbiotica subsp. australis PB68.1, P. temperata subsp. thracensis DSM 15199, P. temperata subsp. temperata M1021 and P. temperata subsp. khani NC19) revealed the presence of up to 22 different BGCs in each of these strains (Tobias et al. 2016b). The main BGCs were NRPS but PKS-NRPS hybrids and other BGC classes were also detected. Interestingly, only a few BGC were found common to all strains: the BGCs for isopropylstilbene (IPS)/Dialkylresorcinol (DAR) and rhabduscin biosynthesis. Rhabduscin (Fig. 4) is an isonitrile NP, a type that is usually rare in nature, and acts as a major player in overcoming the insect immune system via inhibition of the insect enzyme phenoloxidase (Crawford et al. 2012). Phenoloxidase is important in the insect’s blood clotting reaction which involves melanisation and is discussed elsewhere in this volume in the chapter on insect immunity by Eleftherianos et al. So far it is one of the few NP classes found in both Photorhabdus and Xenorhabdus bacteria, as well as in all of their genomes analysed to date (our unpublished data), indicating its important biological function.
DAR with IPS that can be regarded as a special DAR derivative (Fig. 4) and has been shown to be important for development of the nematode symbione (Joyce et al. 2008). Additionally, IPS shows potent antibiotic activity against other bacteria is cytotoxic against eukaryotic cell lines probably due to its inhibition of the soluble epoxide hydrolase (sEH) (Buscató et al. 2013) and DARs have been shown to be signalling compounds involved in quorum sensing in the insect and human pathogen P. asymbiotica (Brameyer et al. 2015) (see also the chapter by Ralf Heermann in this volume). Biochemically, DARs are derived by a consecutive Claisen condensation and Michael addition of a β-keto- and an α,β-unsaturated-acyl thioester, the latter derived from elongation of phenylalanine derived cinnamoyl-CoA, resulting in the formation of a cyclohexanedione (CHD) compound that can be oxidized leading to the formation of DARs (Fig. 5) (Fuchs et al. 2013). When α,β-unsaturated-acyl thioesters from the fatty acid biosynthesis or degradation are used, additional DARs are formed as detected in P. asymbiotica PB68.1. IPS can be further oxidized to give epoxystilbene showing antibiotic and cytotoxic activity and was found in P. luminescens infected larvae of the Greater waxmoth Galleria mellonella (Hu et al. 2006). However, it is also produced when P. luminescens grows in standard Luria Broth (LB) medium and several NPs derived from epoxide opening and additional oxidations have been found (Kontnik et al. 2010). DARs in general seem to be a widespread class of NPs since the corresponding BGCs are found in several different bacteria, several of which are pathogenic to man (Brameyer et al. 2015). Thus, it might be possible that these bacteria also use these NPs as signalling compounds, which would make them a widespread and medically highly relevant NP class that warrants further investigation (Schöner et al. 2015).
Glidobactins (Fig. 4) have been identified from P. asymbiotica infected crickets (Theodore et al. 2012), from P. luminescens grown in low sodium chloride (Stein et al. 2012) and from heterologous expression of the encoding BGC in E. coli (Fu et al. 2012b). The true natural product and major derivative might be cepafungin I, showing an iso-branched acyl moiety. Glidobactin and cepafungin derivatives are potent proteasome inhibitors that could be involved in protecting the insect cadaver against fungi or other invading saprophytes. It’s BGC has been found in all P. luminescens and P. asymbiotica strains examined to date but not in strain DSM 15199 from the P. temperata strains, suggesting that its production may be limited to these groups.
The cyclic pentapeptide GameXPeptides (Bode et al. 2012) have been identified in strain TTO1 and its encoding BGC is found in all but one P. temperata strain examined to date (Nollmann et al. 2015a). In P. luminescens strain TTO1 infected insects derivatives with p-aminophenylalanine (PAPA) or p-methylaminophenylalanine (PMAPA) have been found instead of the usual phenylalanine. Since the titer of these derivatives was much higher, they might actually represent the true natural products produced in a normal infection (Nollmann et al. 2015a). Unfortunately, no biological function has been found for these compounds, despite the fact that they are also widespread in Xenorhabdus strains.
Anthraquinones (AQ) (Li et al. 1995) are responsible for the orange-red pigmentation of P. luminescens and P. temperata and its BGC has been identified in these strains but not in P. asmybiotica which often lacks this orange pigmentation. They are derived from a type II PKS system (Brachmann et al. 2007) that is widespread in Gram-positive Streptomyces bacteria but has so far only been found twice in Gram-negative bacteria. The functions of the AQs are unknown but it has been speculated that they act as ant or bird deterrent factors since similar functions have been observed for AQs in other organisms. However, this has not yet been proven experimentally for AQs from Photorhabdus.
The rhabdopeptide encoding BGC that has been shown in P. luminescens to be responsible for the production of mevalagmapeptides (Bode et al. 2012, 2015a) is present in five of the seven analysed fully sequenced Photorhabdus genomes (see above) and only missing in P. asymbiotica ATCC 43949 and P. temperata NC19. Rhabdopeptides are very common in Xenorhabdus and Photorhabdus strains and some derivatives show both cytotoxic and protozoal activity and thus they might also be involved in protection of the insect cadaver from grazers and saprophytes in the soil. There are a few other BGCs for which no NP has been identified yet, including; two NRPSs, a monomodular type I PKS and additional bacteriocins that are widespread but not common to all strains (Tobias et al. 2016b). Besides these conserved clusters found in several of the analysed strains, additional BGCs are found in only one or two strains.
Kolossin (Fig. 6) is a pentadecapeptide produced by TTO1 (Bode et al. 2015b) that is derived from one of the largest bacterial NRPS with a molecular mass of 1.8 MDa. The Kol NRPS has 46 distinct domains and the respective gene is 49 kbp in length thus alone being 1 % of the bacterial genome. Kolossin could only be obtained from activation of the promoter as discussed below and no function could be assigned yet for this rather simple D/L-peptide made by such a gigantic enzymatic machinery.
Photopyrones (PPY) were found in TTO1 and DSM 15199 (Brachmann et al. 2013) and together with the DARs they are the second example of bacterial quorum sensing molecules different to standard acylhomoserine lactones (AHL) (see also the chapter by Ralf Heermann in this volume). PPYs are made from the same β-keto-thioester also required in DAR and IPS biosynthesis (Fig. 5) but here a homodimeric ketosynthase fuses this thioester to an acyl-thioester resulting in α-pyrone formation (Kresovic et al. 2015). Crucial for the activation of the acylthioester is a glutamine residue in the ketosynthase that enters the active site of the second monomer. Although other α-pyrones are known in NP like corallopyronin or myxopyronine the ketosynthases involved in their biosynthesis do not require the glutamine activation (Erol et al. 2010; Sucipto et al. 2013).
The ink blue coloured pigment indigoidine (5,5′-diamino-4,4′-dihydroxy-3,3′-diazadiphenoquinone-(2,2′)) is derived from a monomodular NRPS found in half of the analysed strains. The compound itself was identified in P. luminescens strain TTO1 using a promoter exchange approach (see below) but no indigoidine has been detected under any growth condition tested (Brachmann et al. 2012). However, from the biophysical properties of indigoidine one might assume a protective function against reactive oxygen species (ROS) or UV light.
Phurealipids have been found in several P. luminescens strains, rarely in P. temperata and Xenorhabdus but are absent from P. asymbiotica (Nollmann et al. 2015b). They have been identified as inhibitors of the juvenile hormone epoxide hydrolase that is required for normal development (moulting) and immunity in insects. Phurealipids are derived from the fatty acid metabolism where they are reduced as aldehydes that are transaminated, carbamoylated and methylated. A carbamoyltransferase and a methyltransferase have been identified in the P. luminscens strain TTO1 genome but a candidate for the aminotransferase is still missing.
Ciche and co-workers isolated a catecholate siderophore, named photobactin, from P. luminescens that is also derived from a NRPS pathway. Due to its role as a siderophore, sequestering and transferring Fe3+ into the bacterial cells, photobactin was required for growth under ion-limited conditions (Ciche et al. 2003). However, photobactin is not needed for P. luminescens to support the growth and the reproduction of its nematode host. However, purified photobactin was shown to have antibiotic activity, suggesting it may play a role in inhibiting competing bacteria in the insect cadaver. Siderophore encoding BGCs are found in all sequenced Photorhabdus strains most of them containing NRPS independent biosynthesis pathways.
In 2004, Ji et al. identified benzylideneacetone (Fig. 6) from X. nematophila being active against some Gram-negative bacteria (Ji et al. 2004). From its structure it might be derived from a cinnamic acid extended by one malonyl-CoA followed by decarboxylation. However, this might suggest the presence of a phenylalanine ammonium lyase and CoA-ligase in X. nematophila that has not been identified in the available genome sequences so far (our unpublished data). In addition to benzylideneacetone, a linear proline-tyrosine dipeptide and an acetylated phenylalanine-glycine-valine tripeptide were both isolated and they were also shown to be phospholipase A2 inhibitors. These three compounds could also be isolated from P. temperata culture broth. Additionally, X. nematophila can produce four additional phospholipase A2 inhibitors (indole, oxidole, cyclo-proline-tyrosine dipeptide and p-hydroxyphenyl propionic acid) (Seo et al. 2012). Phospholipase A2 is crucial for the insect’s immune response and thus needs to be overcome in order to colonize the insect.
Two very small compounds, benzaldehyde (Ullah et al. 2015) and phthalic acid (Ullah et al. 2014), were isolated from P. temperata. Benzaldehyde possessed antioxidant, insecticidal and antimicrobial activities. Moreover, phthalic acid has the capacity to inhibit phenoloxidase with a consequence to suppress the insect’s immune defence.
Derzelle and co-workers have identified the putative gene cluster of a carbapenem-like antibiotic in P. luminescens (Derzelle et al. 2002) and two types of carbapenem BGCs have been identified in four of the seven genomes annotated to date. However, their exact structure is not known. Carbapenem antibiotics are members of the β-lactam family of antibiotics, which are now the most important class of antibiotics for clinical use and their biosynthesis and regulation has been extensively studied in Serratia (Coulthurst et al. 2005).
4 Tools for the Identification of Natural Products
In order to identify NPs from Photorhabdus or any other organism one can cultivate the strain of interest using several different growth conditions summarized in the OSMAC (One Strain Many Compounds) approach (Bode et al. 2002) that often results in the production of different NP classes at different conditions varying media composition, aeration, pH, salt concentration and even co-cultivation with other organisms to mimic the natural condition at which the respective NP is produced. One can also try a range of molecular techniques to enhance the production of the NP in order to enhance its detection and purification. Each of these will now be discussed below.
4.1 Promoter Exchange
One can also use the BGC information obtained from whole genome sequencing for a ‘promoter exchange’ approach. Since the natural promoters might have unknown modes of regulation via specific transcription factors or other unknown mechanisms, they can be replaced by strong constitutive or inducible promoters (Biggins et al. 2014, 2011; Bode et al. 2015a). The latter approach is especially beneficial since it allows to compare the wild-type strain with the induced (usually overproduction of the NP derived from BGC overexpression) and the non-induced promoter exchange mutant (usually a non-producer due to the missing expression) (Fig. 7). This simple approach has also been used widely by others but was especially useful in Photorhabdus and Xenorhabdus with the arabinose inducible P BAD/AraC system resulting in the production of xenoamicins, GameXPeptides, mevalagmapeptides, the blue pigment indigoidine, the cytotoxic yellow pigment xenorhabdin and kolossin (Bode et al. 2015a, b). The latter is derived from one of the largest bacterial NRPS, named Kol, with a molecular weight of 1.8 MDa and 49 catalytically active domains covering almost 1 % of the Photorhabdus chromosome. Kol is only expressed in a promoter exchange mutant and no natural condition has been found so far for its production (Bode et al. 2015b). The power of the method is also evident from the fact that for xenoamicin productin in Xenorhabdus doucetiae the production titer is well beyond the solubility of these depsipeptides and white crystals can therefore be observed in the expression culture. For GameXPeptides the 10-fold higher production titer compared to the wild-type level was accompanied by the occurrence of the linear versions of the usually cyclic peptides (Bode et al. 2015a).
Schematic representation of the promoter exchange approach used in Photorhabdus and Xenorhabdus (Bode et al. 2015a). The start (300–600 bp) of the gene of interest is amplified by PCR and cloned into the cluster expression plasmid pCEP. Following transformation into Photorhabdus, the genomic insertion of the non-replicating plasmid results in a promoter exchange mutant in which the expression of the full-length gene is not driven by the natural promoter (grey arrow) but by the introduced inducible promoter (green arrow; here P BAD). The resulting promoter exchange mutant can easily be selected based on the pCEP-encoded resistance gene. The new promoter is tightly controlled and shows no activity without the inducer arabinose. Therefore, the non-induced strain behaves like a knockout mutant (no production of the compound of interest) whereas overexpression of the desired gene is achieved with arabinose resulting in an overproducing mutant (in green, relative to the wild-type strain). From the comparison of these three strains the NP can easily correlated to the BGC in the genome
4.2 Manipulation of Regulatory Proteins
Another approach is the manipulation of regulatory proteins involved in NP production. This approach was especially useful for fungi that often encode a specific transcriptional regulator as part of their NP BGC (Scharf and Brakhage 2013). In Photorhabdus it was shown that HexA (see chapter by David Clarke also in this volume) and the two-component system BarA-UvrY (see chapter by David Clarke in this volume) are involved in NP regulation. Previous research has concentrated on the UV active NP isopropylstilbene and anthraquinones that are both influenced by these regulators. A detailed analysis of a hexA deletion and overexpression mutant using the promoter exchange approach described above revealed that in a hexA deletion GameXPeptides and mevalagmapeptides are overproduced while some phurealipids are strongly decreased (our unpublished data). Similar analyses with the global regulators LeuO and Lrp showed that they also affect NP production. LeuO is a LysR type transcription factor (Hernandez-Lucas and Calva 2012) involved in virulence of Vibrio cholerae and Lrp (Brinkman et al. 2003) has been described as a global regulator affecting mutualism with the nematode and pathogenicity against the insect in Xenorhabdus nematophila (Cowles et al. 2007). We were able to show that Lrp is required for the production of rhabdopeptides in X. nematophila (our unpublished data) that show insecticidal activity. In P. luminescens TT01 lrp deletion increases mevalagmapeptide production but decreases phurealipid production while overexpression restored wild-type production levels but led to increased desmethylphurealipid levels. Phurealipid biosynthesis is not encoded in an operon and currently only two steps in their biosynthesis encoded in different genomic loci are known. However, from these results it is quite obvious that both genes are differentially regulated leading to the observed production phenotype using hexA, lrp or leuO mutants.
The most dramatic effect of all regulatory mutants analysed in our group was that of a hfq deletion mutant in P. luminescens. Hfq is a RNA chaperone mediating interaction between small regulatory RNA and mRNA that is involved in the regulation of virulence in several different bacteria (De Lay et al. 2013; Vogel and Luisi 2011). In P. luminescens a ∆hfq strain did not produce any known natural product (Fig. 8). In fact the strain looked more like an E. coli strain with essentially no NP produced whereas genetic complementation with a plasmid encoded hfq restored NP production although not to the wild-type level for all NP classes (Tobias et al. 2016a). Interestingly, while no decrease in insect pathogenicity was observed for a ∆hfq strain, no nematode recovery at all was observed when Heterorhabditis bacteriophora was developed on the ∆hfq strain. This might indicate no crucial role of NP in insect pathogenesis but an even more important function in the symbiosis with the nematode as suggested previously for isopropylstilbene (Joyce et al. 2008). Here the next step clearly is the identification of the underlying regulatory principles and especially the small RNA(s) that cause this severe phenotype.
4.3 Heterologous Expression
In general heterologous production of BGCs from Photorhabdus and Xenorhabdus works well in E. coli and direct cloning and yeast based cloning have both applied successfully in the past (Crawford et al. 2012; Fu et al. 2012a; Schimming et al. 2014). However, the promoter exchange approach as well as the manipulation of regulatory proteins might be superior to the heterologous expression of the BGC of interest since all precursors needed for the production of the ‘real’ NP are present in the original producer while the heterologous host might not have these precursors. Even simple building blocks like amino or fatty acids can be different and for P. luminescens it has been shown that heterologous expression of the GameXPeptide producing NRPS GxpS in E. coli led to the production of the usual GameXPeptides A-D while overexpression of gxpS results in derivatives carrying p-amino- or p-aminomethyl phenylalanine (Nollmann et al. 2015a). These derivatives are the major derivatives produced in insects since this activates a BGC for the production of these unusual phenylalanine derivatives. Similarly, heterologous expression of the glidobactin producing BGC resulted in the production of glidobactin A in E. coli (Dudnik et al. 2013; Fu et al. 2012a) while the major derivative in P. luminescens is in fact cepafungin (Stein et al. 2012; Theodore et al. 2012) having an iso-branched fatty acid. However, E. coli is not able to produce such fatty acids.
4.4 Chemical Tools
Beyond these molecular methods, one can also use further analytical chemistry to identify and characterize NP. Here the major breakthrough was the development of mass spectrometry to allow the very sensitive detection of NPs and even the visualization of NP production in symbiotic systems or other interactions. Mass spectrometry also allows the structural elucidation of NPs without their actual isolation and in combination with labelling experiments even the configuration of a peptide NP can be assigned as demonstrated for the GameXPeptides and kolossin from Photorhabdus (Bode et al. 2012, 2015b).
Isotopic labelling can be regarded as a special form of precursor-directed biosynthesis that allows the identification of NP building blocks and even the generation of NP derivatives with modified biological activity. Even more useful would be the potential incorporation of a chemical label that not only allows for the rapid localization of any given NP but can be also used for NP enrichment. One such label is the azide and/or alkyne group as these groups are small and are often well tolerated by the biosynthesis machinery of the cell. They can also react efficiently and specifically without complicated workup in a Huisgen 1,3-dipolar cycloaddition (Kolb et al. 2001). The proven applications of this prototype of a so-called ‘click reaction’ range from inorganic and organic synthesis to in vitro labelling. The true milestone in development and optimization of this technique was achieved by Bertozzi and co-workers via the rediscovery of ring-strained cyclooctynes as reaction partners for azides without the need of any possibly cytotoxic copper catalysts, a process coined the strain-promoted azide-alkyne cycloaddition (Fig. 9) (Jewett and Bertozzi 2010).
This discovery opened up the now very popular field of ‘bioorthogonal’ chemistry, where two reaction partners (e.g. an alkyne and a cyclooctyne) can react even within a live cell without substantially altering or harming it. This is possible because in most organisms there is no natural reaction partner for either a cyclooctyne or an azide is present. With the azido group also being a rather small functional group, roughly the size of an ethyl group, not featuring a noteworthy polarity or reactivity, it is the perfect label for in vivo investigations.
Since fatty acids with ω-alkyne or –azide groups have been used previously for the detection and enrichment of lipoproteins (Hang and Linder 2011; Hang et al. 2007), we tested the incorporation of ω-azide fatty acids (AFA) in NP produced by Photorhabdus and Xenorhabdus and indeed we could detect such derivatives after reaction with an cleavable azide reactive resin (CARR) and the subsequent enrichment of triazole (Fig. 10) (Pérez et al. 2016). The advantage of this method is the gain in MS sensitivity through the very efficient ionization of the triazole product formed as well as the characteristic fragmentation pattern of the clicked derivative that enabled the reliable identification of the clicked products and thus the original azide containing NP.
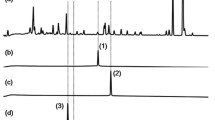
Overview on azide enrichment using cleavable azide reactive resin (CARR) showing the typical fragmentation of triazole products formed as well as a typical example from a bacterial extract before and after CARR enrichment indicating the purification of only CARR-reacted azides (see BPC and EIC for comparison). BPC (base peak chromatogram = all ions are shown), EIC (extracted ion chromatogram = only ion traces for specific compounds are shown), TCEP (tris(2-carboxyethyl)phosphine))
In Photorhabdus AFA-based CARR led to the identification of phurealipids, whereas in X. doucetiae phenylethylamides were detected. Applying p-azidophenylalanine based CARR in P. luminescens GameXPeptide A, cinnamic acid and cinnamoyl-phenylalanine (all in azidated form) have been identified (Pérez et al. 2016). The latter represents a new class of NP from P. luminescens of which the non-azidated form has not been identified yet. Interestingly, similar compounds were also identified in X. szentirmaii during p-azidophenylalanine-based CARR (Fig. 11).
The fact that also azidated peptides like GameXPeptide or szentiamide from X. szentirmaii or xenortide from X. nematophila have been detected resulting from the substitution of phenylalanine by p-azidophenylalanine indicates the flexibility of the responsible NRPS to incorporate even these non-natural amino acids. Such peptides might be of great interest for the drug discovery process since they can be further modified chemically after their isolation using the same click chemistry used in the CARR enrichment.
5 Conclusions
During the last 10 years NP research within entomopathogenic bacteria has made huge advances due to the diligent work of several laboratories. However, there is still a lot of information missing that requires further research. One goal for the future must be to develop more tools for the rapid target identification of the various NPs produced. We need these tools to answer the wide range outstanding biological questions that remain. For example, to what enzyme or receptor do these NPs bind and what is the resulting phenotype in the insect, the nematode, the original producer or food competitors? To achieve this goal classical mode of action studies must be performed (Schmitt et al. 2015), however, these are often long and technically challenging studies. Reactive NP derivatives can be synthesized that can covalently bind to the NP target and in case a handle like an azide is also present in these NP derivatives one can use this to isolate the NP-target complex as previously shown.
Additionally, if one assumes that all Photorhabdus strains are adapted to the same ecological niche being mutualistic to nematodes and pathogenic towards insects. Thus, they might require a certain set of functionally similar chemical tools to fulfil these similar ecological functions. Therefore, one might predict NPs that are functionally similar but can be chemically different and might be derived from different BGCs in strains that do not have a NP with a known functionality. Examples of this strain specificity of NP production might be the P. temperata strains lacking glidobactins. In which case, here it would be interesting to look for other proteasome inhibitors that might have chemically new scaffolds.
Regarding proteasome inhibitors, and other toxic NP compounds, it is still a mystery why the nematode host can survive in the dead insect containing such large amounts of these toxic compounds. Once the targets of these NP in the insect are known one must investigate whether they are different in the nematode host or how it can protect itself from their activity. A resistance against toxic NPs from the bacterial symbiont can also be a protection mechanism against other nematodes and might ensure the specificity of the symbiosis by killing the wrong nematode.
Finally, with the already developed molecular tools for manipulation of the bacteria and the first available genome sequences for the entomopathogenic nematodes (Bai et al. 2013; Dillman et al. 2015) that allow also transcriptomics and proteomics one can also start to address the NP function from the hosts side by manipulation of NP regulated signalling pathways or other NP targets. Moreover, similar work can be done in insects where all tools are already available and just need to be used to address the functions of NPs from entomopathogenic bacteria that again can be analysed in depth using ‘omics’ technologies. It is an exciting time to study these bacteria and once we have understood such a rather simple system of organismic interaction we can also start to look in detail in other and more complex systems involving more interaction partners.
References
Ackermann M (2015) A functional perspective on phenotypic heterogeneity in microorganisms. Nat Publishing Group 13:497–508
Arnison PG, Bibb MJ, Bierbaum G, Bowers AA, Bugni TS, Bulaj G, Camarero JA, Campopiano DJ, Challis GL, Clardy J et al (2012) Ribosomally synthesized and post-translationally modified peptide natural products: overview and recommendations for a universal nomenclature. Nat Prod Rep 30:108–160
Bai X, Adams BJ, Ciche TA, Clifton S, Gaugler R, Kim K-S, Spieth J, Sternberg PW, Wilson RK, Grewal PS (2013) A lover and a fighter: the genome sequence of an entomopathogenic nematode heterorhabditis bacteriophora. PLoS One 8:e69618
Biggins JB, Liu X, Feng Z, Brady SF (2011) Metabolites from the induced expression of cryptic single operons found in the genome of Burkholderia pseudomallei. J Am Chem Soc 133:1638–1641
Biggins JB, Kang H-S, Ternei MA, DeShazer D, Brady SF (2014) The chemical arsenal of Burkholderia pseudomalleiIs essential for pathogenicity. J Am Chem Soc 136:9484–9490
Bode HB, Müller R (2005) The impact of bacterial genomics on natural product research. Angew Chem Int Ed 44:6828–6846
Bode HB, Bethe B, Höfs R, Zeeck A (2002) Big effects from small changes: possible ways to explore nature’s chemical diversity. ChemBioChem 3:619–627
Bode HB, Reimer D, Fuchs SW, Kirchner F, Dauth C, Kegler C, Lorenzen W, Brachmann AO, Grün P (2012) Determination of the absolute configuration of peptide natural products by using stable isotope labeling and mass spectrometry. Chem Eur J 18:2342–2348
Bode E, Brachmann AO, Kegler C, Simsek R, Dauth C, Zhou Q, Kaiser M, Klemmt P, Bode HB (2015a) Simple “on-demand” production of bioactive natural products. ChemBioChem 16:1115–1119
Bode HB, Brachmann AO, Jadhav KB, Seyfarth L, Dauth C, Fuchs SW, Kaiser M, Waterfield NR, Sack H, Heinemann SH et al (2015b) Structure elucidation and activity of kolossin A, the D-/L-Pentadecapeptide product of a giant nonribosomal peptide synthetase. Angew Chem Int Ed Engl 54:10352–10355
Brachmann AO, Joyce SA, Jenke-Kodama H, Schwär G, Clarke DJ, Bode HB (2007) A type II polyketide synthase is responsible for anthraquinone biosynthesis inphotorhabdus luminescens. ChemBioChem 8:1721–1728
Brachmann AO, Kirchner F, Kegler C, Kinski SC, Schmitt I, Bode HB (2012) Triggering the production of the cryptic blue pigment indigoidine from Photorhabdus luminescens. J Biotechnol 157:96–99
Brachmann AO, Brameyer S, Kresovic D, Hitkova I, Kopp Y, Manske C, Schubert K, Bode HB, Heermann R (2013) Pyrones as bacterial signaling molecules. Nat Chem Biol 9:573–578
Brameyer S, Kresovic D, Bode HB, Heermann R (2015) Dialkylresorcinols as bacterial signaling molecules. Proc Natl Acad Sci U.S.A. 112:572–577
Brinkman AB, Ettema TJG, De Vos WM, Van Der Oost J (2003) The Lrp family of transcriptional regulators. Mol Microbiol 48:287–294
Buscató EL, Büttner D, Brüggerhoff A, Klingler F-M, Weber J, Scholz B, Živković A, Marschalek R, Stark H, Steinhilber D et al (2013) From a multipotent stilbene to soluble epoxide hydrolase inhibitors with antiproliferative properties. ChemMedChem 8:919–923
Caboche S, Leclere V, Pupin M, Kucherov G, Jacques P (2010) Diversity of monomers in nonribosomal peptides: towards the prediction of origin and biological activity. J Bacteriol 192:5143–5150
Cantley AM, Clardy J (2015) Animals in a bacterial world: opportunities for chemical ecology. Nat Prod Rep 32:888–892
Chiocchini C, Linne U, Stachelhaus T (2006) In vivo biocombinatorial synthesis of lipopeptides by COM domain-mediated reprogramming of the surfactin biosynthetic complex. Chem Biol 13:899–908
Ciche TA, Blackburn M, Carney JR, Ensign JC (2003) Photobactin: a Catechol Siderophore Produced by Photorhabdus luminescens, an Entomopathogen Mutually Associated with Heterorhabditis bacteriophora NC1 Nematodes. Appl Environ Microbiol 69:4706–4713
Cosmina P, Rodriguez F, de Ferra F, Grandi G, Perego M, Venema G, van Sinderen D (1993) Sequence and analysis of the genetic locus responsible for surfactin synthesis in Bacillus subtilis. Mol Microbiol 8:821–831
Coulthurst SJ, Barnard AML, Salmond GPC (2005) Regulation and biosynthesis of carbapenem antibiotics in bacteria. Nat Rev Micro 3:295–306
Cowles KN, Cowles CE, Richards GR, Martens EC, Goodrich-Blair H (2007) The global regulator Lrp contributes to mutualism, pathogenesis and phenotypic variation in the bacterium Xenorhabdus nematophila. Cell Microbiol 9:1311–1323
Crawford JM, Portmann C, Zhang X, Roeffaers MBJ, Clardy J (2012) Small molecule perimeter defense in entomopathogenic bacteria. Proc Natl Acad Sci U.S.A. 109:10821–10826
Davies J, Ryan KS (2012) Introducing the parvome: bioactive compounds in the microbial world. ACS Chem Biol 7:252–259
Davies J, Spiegelman GB, Yim G (2006) The world of subinhibitory antibiotic concentrations. Curr Opin Microbiol 9:445–453
De Lay N, Schu DJ, Gottesman S (2013) Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288:7996–8003
Derzelle S, Duchaud E, Kunst F, Danchin A, Bertin P (2002) Identification, characterization, and regulation of a cluster of genes involved in carbapenem biosynthesis in Photorhabdus luminescens. Appl Environ Microbiol 68:3780–3789
Dillman AR, Macchietto M, Porter CF, Rogers A, Williams B, Antoshechkin I, Lee M-M, Goodwin Z, Lu X, Lewis EE et al (2015) Comparative genomics of steinernema reveals deeply conserved gene regulatory networks. Genome Biol 1–21
Dorrestein PC, Mazmanian SK, Knight R (2014) Finding the missing links among metabolites, microbes, and the host. Immunity 40:824–832
Dudnik A, Bigler L, Dudler R (2013) Heterologous expression of a Photorhabdus luminescens syrbactin-like gene cluster results in production of the potent proteasome inhibitor glidobactin A. Microbiol Res 168:73–76
Erol Ö, Schäberle TF, Schmitz A, Rachid S, Gurgui C, Omari, EM, Lohr F, Kehraus S, Piel J, Müller R et al (2010) Biosynthesis of the myxobacterial antibiotic corallopyronin A. ChemBioChem 11:1253–1265
Felnagle EA, Jackson EE, Chan YA, Podevels AM, Berti AD, McMahon MD, Thomas MG (2008) Nonribosomal peptide synthetases involved in the production of medically relevant natural products. Mol Pharm 5:191–211
Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Müller R, Stewart AF et al (2012a) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446
Fu J, Bian X, Hu S, Wang H, Huang F, Seibert PM, Plaza A, Xia L, Stewart AF, ller RMU et al (2012b) Full-length RecE enhances linear-linear homologous recombination and facilitates direct cloning for bioprospecting. Nat Biotechnol 30:440–446
Fuchs SW, Bozhüyük KAJ, Kresovic D, Grundmann F, Dill V, Brachmann AO, Waterfield NR, Bode HB (2013) Formation of 1,3-cyclohexanediones and resorcinols catalyzed by a widely occurring ketosynthase. Angew Chem Int Ed 52:4108–4112
Gokhale RS, Khosla C (2000) Role of linkers in communication between protein modules. Curr Opin Chem Biol 4:22–27
Grünewald J, Marahiel MA (2006) Chemoenzymatic and template-directed synthesis of bioactive macrocyclic peptides. Microbiol Mol Biol Rev 70:121–146
Gulick AM (2009) Conformational dynamics in the Acyl-CoA synthetases, adenylation domains of non-ribosomal peptide synthetases, and firefly luciferase. ACS Chem Biol 4:811–827
Hahn M, Stachelhaus T (2004) Selective interaction between nonribosomal peptide synthetases is facilitated by short communication-mediating domains. Proc Natl Acad Sci 101:15585–15590
Hahn M, Stachelhaus T (2006) Harnessing the potential of communication-mediating domains for the biocombinatorial synthesis of nonribosomal peptides. Proc Natl Acad Sci 103:275–280
Hang HC, Linder ME (2011) Exploring protein lipidation with chemical biology. Chem Rev 111:6341–6358
Hang HC, Geutjes E-J, Grotenbreg G, Pollington AM, Bijlmakers MJ, Ploegh HL (2007) Chemical probes for the rapid detection of fatty-acylated proteins in mammalian cells. J Am Chem Soc 129:2744–2745
Hernandez-Lucas I, Calva E (2012) The coming of age of the LeuO regulator. Mol Microbiol 85:1026–1028
Hu K, Li J, Li B, Webster JM, Chen G (2006) A novel antimicrobial epoxide isolated from larval Galleria mellonella infected by the nematode symbiont, Photorhabdus luminescens (Enterobacteriaceae). Bioorg Med Chem 14:4677–4681
Ikeda H, Ishikawa J, Hanamoto A, Shinose M, Kikuchi H, Shiba T, Sakaki Y, Hattori M, Ōmura S (2003) Complete genome sequence and comparative analysis of the industrial microorganism streptomyces avermitilis. Nat Biotechnol 21:526–531
Jewett JC, Bertozzi CR (2010) Cu-free click cycloaddition reactions in chemical biology. Chem Soc Rev 39:1272–1279
Ji D, Yi Y, Kang G-H, Choi Y-H, Kim P, Baek N-I, Kim Y (2004) Identification of an antibacterial compound, benzylideneacetone, from Xenorhabdus nematophilaagainst major plant-pathogenic bacteria. FEMS Microbiol Lett 239:241–248
Joyce SA, Brachmann AO, Glazer I, Lango L, Schwär G, Clarke DJ, Bode HB (2008) Bacterial biosynthesis of a multipotent stilbene. Angew Chem Int Ed 47:1942–1945
Kolb HC, Finn MG, Sharpless KB (2001) Click-Chemie: diverse chemische Funktionalität mit einer Handvoll guter Reaktionen. Angew Chem 113:2056–2075
Kontnik R, Crawford JM, Clardy J (2010) Exploiting a global regulator for small molecule discovery in Photorhabdus luminescens. ACS Chem Biol 5:659–665
Kresovic D, Schempp F, Cheikh-Ali Z, Bode HB (2015) A novel and widespread class of ketosynthase is responsible for the head-to-head condensation of two acyl moieties in bacterial pyrone biosynthesis. Beilstein J Org Chem 11:1412–1417
Lautru S, Challis GL (2004) Microbiology 150(Pt 6):1629–1636. Review. PMID: 15184549
Li J, Chen G, Wu H, Webster JM (1995) Identification of two pigments and a hydroxystilbene antibiotic from Photorhabdus luminescens. Appl Environ Microbiol 61:4329–4333
Ling LL, Schneider T, Peoples AJ, Spoering AL, Engels I, Conlon BP, Mueller A, Schäberle TF, Hughes DE, Epstein S et al (2015) A new antibiotic kills pathogens without detectable resistance. Nature 517:455–459
Marahiel MA (2016) A structural model for multimodular NRPS assembly lines. Nat Prod Rep 33:136–140
Mootz HD, Marahiel MA (1997) The tyrocidine biosynthesis operon of Bacillus brevis: complete nucleotide sequence and biochemical characterization of functional internal adenylation domains. J Bacteriol 179:6843–6850
Mootz HD, Schwarzer D, Marahiel MA (2002) Ways of assembling complex natural products on modular nonribosomal peptide synthetases. ChemBioChem 3:490–504
Newman DJ, Cragg GM (2012) Natural products as sources of new drugs over the 30 years from 1981 to 2010. J Nat Prod 75:311–335
Nollmann FI, Dauth C, Mulley G, Kegler C, Kaiser M, Waterfield NR, Bode HB (2015a) Insect-specific production of new GameXPeptides in Photorhabdus luminescens TTO1, widespread natural products in entomopathogenic bacteria. ChemBioChem 16:205–208
Nollmann FI, Heinrich AK, Brachmann AO, Morisseau C, Mukherjee K, Casanova-Torres ÁM, Strobl F, Kleinhans D, Kinski S, Schultz K et al (2015b) A Photorhabdus natural product inhibits insect juvenile hormone epoxide hydrolase. ChemBioChem 16:766–771
Omura S, Ikeda H, Ishikawa J, Hanamoto A, Takahashi C, Shinose M, Takahashi Y, Horikawa H, Nakazawa H, Osonoe T et al (2001) Genome sequence of an industrial microorganism streptomyces avermitilis: deducing the ability of producing secondary metabolites. Proc Natl Acad Sci 98:12215–12220
Pérez AJ, Wesche F, Adihou H, Bode HB (2016) Solid-phase enrichment and analysis of azide-labeled natural products: fishing downstream of biochemical pathways. Chemistry 22:639–645
Scharf DH, Brakhage AA (2013) Engineering fungal secondary metabolism: a roadmap to novel compounds. J Biotechnol 163:179–183
Schimming O, Fleischhacker F, Nollmann FI, Bode HB (2014) Yeast homologous recombination cloning leading to the novel peptides ambactin and xenolindicin. ChemBioChem 15:1290–1294
Schmitt EK, Hoepfner D, Krastel P (2015) Natural products as probes in pharmaceutical research. J Ind Microbiol Biotechnol 43:249–260
Schöner TA, Kresovic D, Bode HB (2015) Biosynthesis and function of bacterial dialkylresorcinol compounds. Appl Microbiol Biotechnol 1–6
Seo S, Lee S, Hong Y, Kim Y (2012) Phospholipase A2 inhibitors synthesized by two entomopathogenic bacteria, xenorhabdus nematophila and Photorhabdus temperata subsp. temperata. Appl Environ Microbiol 78:3816–3823
Sharon G, Garg N, Debelius J, Knight R, Dorrestein PC, Mazmanian SK (2014) Specialized metabolites from the microbiome in health and disease. Cell Metab 20:719–730
Sieber SA, Marahiel MA (2005) Molecular mechanisms underlying nonribosomal peptide synthesis: approaches to new antibiotics. Chem Rev 105:715–738
Stein ML, Beck P, Kaiser M, Dudler R (2012) One-shot NMR analysis of microbial secretions identifies highly potent proteasome inhibitor
Sucipto H, Wenzel SC, Müller R (2013) Exploring chemical diversity of α-Pyrone antibiotics: molecular basis of myxopyronin biosynthesis. ChemBioChem 14:1581–1589
Theodore CM, King JB, You J, Cichewicz RH (2012) Production of cytotoxic glidobactins/luminmycins by Photorhabdus asymbioticain liquid media and live crickets. J Nat Prod 75:2007–2011
Tobias NJ, Heinrich AK, Eresmann H, Wright PR, Neubacher N, Backofen R, Bode HB (2016a) Environ Microbiol. Aug 23. doi:10.1111/1462-2920.13502
Tobias NJ, Mishra B, Gupta DK, Sharma R, Thines M, Stinear TP, Bode HB (2016b) Genome comparisons provide insights into the role of secondary metabolites in the pathogenic phase of the Photorhabdus life cycle. BMC Genomics
Ullah I, Khan A, Ali L, Khan A, Waqas M, Lee I-J, Shin J-H (2014) An Insecticidal compound produced by an insect-pathogenic bacterium suppresses host defenses through phenoloxidase inhibition. Molecules 19:20913–20928
Ullah I, Khan AL, Ali L, Khan AR, Waqas M, Hussain J, Lee I-J, Shin J-H (2015) Benzaldehyde as an insecticidal, antimicrobial, and antioxidant compound produced by Photorhabdus temperata M1021. J Microbiol 53:127–133
Vogel J, Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Micro 9:578–589
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2016 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Bozhüyük, K.A.J., Zhou, Q., Engel, Y., Heinrich, A., Pérez, A., Bode, H.B. (2016). Natural Products from Photorhabdus and Other Entomopathogenic Bacteria. In: ffrench-Constant, R. (eds) The Molecular Biology of Photorhabdus Bacteria . Current Topics in Microbiology and Immunology, vol 402. Springer, Cham. https://doi.org/10.1007/82_2016_24
Download citation
DOI: https://doi.org/10.1007/82_2016_24
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-52714-7
Online ISBN: 978-3-319-52715-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)