Abstract
The transfer of ADP-ribose from NAD+ to a substrate by ADP-ribosyltransferases, ADP-ribosylation, is a multifunctional posttranslational modification. While many studies have addressed the function of poly-ADP-ribosylation, for example, in DNA repair, signaling, and gene transcription, little is known about the role of mono-ADP-ribosylation. Recent work describing the mono-ADP-ribosyltransferase ARTD10/PARP10 suggests that this enzyme affects apoptosis, NF-κB signaling, and DNA damage repair, at least in part dependent on its activity as mono-ADP-ribosyltransferase. Moreover, the macrodomain-containing proteins MacroD1, MacroD2, and TARG1/C6orf130 were recently described as hydrolases, which remove mono-ADP-ribosylation thus providing evidence that this modification is reversible. In this review, we discuss these novel findings and their broader implications for cell behavior. We suggest functions of ARTD10 in immunity, metabolism, and cancer biology.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Newcastle Disease Virus
- Nuclear Localization Sequence
- Venezuelan Equine Encephalitis Virus
- APOB Gene
- Venezuelan Equine Encephalitis
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
Controlling the activities of proteins transiently and independently of gene transcription is fundamental for the regulation of numerous cellular processes. This is achieved by modulating the function of proteins by post-translational modifications (PTMs). A large number of PTMs exist, including phosphorylation, glycosylation, and ubiquitination, which can affect, for example, the subcellular localization, the catalytic activity, or the half-life of the modified protein. ADP-ribosylation is a PTM, where ADP-ribose is linked to proteins either as a monomer––called mono-ADP-ribosylation––or where chains of multiple ADP-ribose units are formed, which is referred to as poly-ADP-ribosylation. The addition of a single ADP-ribose moiety introduces negative charges to the modified substrate and thus is thought to affect protein function. Poly-ADP-ribosylation creates linear or branched ADP-ribose chains, which serve as scaffolds for other proteins to interact with. For instance, DNA repair proteins and transcriptional cofactors and cofactor complexes have been shown to be recruited by poly-ADP-ribose (De Vos et al. 2012; Gibson and Kraus 2012; Kleine and Luscher 2009; Schreiber et al. 2006). The length of the polymers suggests that multiple proteins may bind simultaneously and thus poly-ADP-ribose may promote the assembly of large protein complexes. Poly-ADP-ribosylation may also directly affect protein function, for example the automodification of ARTD1 in response to DNA damage inhibits eventually DNA binding (D’Amours et al. 1999; Kraus 2008). Currently 22 ADP-ribosyltransfereases have been described in humans. These proteins can be subdivided into two families according to their homology to bacterial toxins, which also resulted in the suggestion for an updated nomenclature (Hottiger et al. 2010). The ARTC family of ADP-ribosyltransferases are found at the cell membrane and ADP-ribosylate arginine residues (Laing et al. 2011). The residues involved in NAD+ binding and catalysis, Arg-Ser-Glu, are also present in certain toxins such as cholera toxin, hence the name ADP-ribosyltransferases Cholera toxin-like. Intracellular mono-ADP-ribosylation and poly-ADP-ribosylation are catalyzed by the ADP-ribosyltransferase Diphtheria toxin-like family (ARTDs), also referred to as the PARP family (Fig. 1), which have a distinct amino acid motif, His-Tyr-Glu, compared to ARTCs that is key to execute ADP-ribosylation (Hottiger et al. 2010). The ARTD family can be further subdivided into proteins with iterative mono-ADP-ribosyltransferase activity and thus capable to form ADP-ribose polymers, proteins with single mono-ADP-ribosyltransferase activity, and inactive family members (Hottiger et al. 2010). These three subfamilies are characterized with distinct alterations in the amino acid motif. In particular the replacement of the catalytically relevant Glu distinguishes polymer-forming enzymes from mono-ADP-ribosyltransferases (Fig. 1). The distinction between mono- and poly-ADP-ribosylation is further emphasized by the existence of both mono-ADP-ribose––and poly-ADP-ribose-specific reader domains and hydrolases, which recognize and reverse these modifications (Barkauskaite et al. 2013; Feijs et al. 2013a).
Summary of the catalytic activities of intracellular ADP-ribosyltransferase (ARTD) family members. ADP-ribosylation involves the transfer of ADP-ribose from NAD+ to a substrate and release of nicotinamide (NAM). The ARTD family enzymes are subdivided according to differences in their catalytic activity. The activity of poly-ADP-ribosyltransferases such as ARTD1 depends on three critical amino acid residues (His-Tyr-Glu) in the active center/cofactor binding site, two of which (His and Tyr) are important to interact with NAD+, while the glutamate stabilizes an oxacarbenium intermediate during catalysis. These amino acids, which are referred to as the catalytic motif, are not conserved in all ARTDs. As a consequence the catalytic activities are changed. Hence three different subfamilies of ARTDs have been proposed. Class I ARTDs are poly-ADP-ribosyltransferases with the original motif HYE. In class II ARTDs, including ARTD10, the acidic glutamate is replaced by I/L/T/V/Y, which results in restriction to mono-ADP-ribosylation. This class of enzymes uses substrate-assisted catalysis to modify substrates, i.e., a glutamate of the substrate activates NAD+ but is subsequently modified and cannot be reused for a second reaction. In the third class also the basic histidine residue is replaced with an uncharged amino acid and these enzymes lose the ability to bind NAD+ rendering them catalytically inactive
ARTD1/PARP1 is the best-characterized poly-ADP-ribosyltransferase. It reacts to different forms of cellular stress by its influence on DNA repair, gene transcription, chromatin remodeling, and signal transduction mechanisms, thereby contributing to, for example, an inflammatory response and the survival or death of cells (De Vos et al. 2012; Krishnakumar and Kraus 2010; Luo and Kraus 2012; Messner and Hottiger 2011). ADP-ribosylation of proteins was recognized 50 years ago and a distinction was made between mono-ADP-ribosylation and poly-ADP-ribosylation in cells soon after (Hassa et al. 2006). However, the mono-ADP-ribosyltransferases investigated were predominantly bacterial toxins and extracellular enzymes, while it remained unclear whether intracellular mono-ADP-ribosyltransferases exist (Deng and Barbieri 2008; Di Girolamo et al. 2005). ARTD10/PARP10 was the first intracellular ARTD, which was identified as a bona fide mono-ADP-ribosyltransferase, defining a new subfamily of ADP-ribosyltransferases with nine members (Hottiger et al. 2010; Kleine et al. 2008). The functional characterization of these enzymes is still rather preliminary compared with at least some of the poly-ADP-ribosylating enzymes. However, the last couple of years have seen the beginning of the elucidation of pathways and processes that are affected by mono-ADP-ribosyltransferases (Feijs et al. 2013c). Here we summarize these findings and discuss broader implications of these observations with an outlook towards a potential role of ARTD10 in immunity, metabolism, and cancer biology. Moreover, we highlight open questions that need to be addressed to understand mono-ADP-ribosylation in more general terms.
2 Mechanism of Mono-ADP-Ribosylation by ARTD10
ARTDs catalyze the addition of ADP-ribose to proteins by consuming the co-factor NAD+ and releasing nicotinamid (Fig. 1). The catalytic domain of ARTD10 was identified in a homology search for ADP-ribosyltransferase domains based on the catalytic domain of ARTD1 (Ame et al. 2004; Otto et al. 2005). Later, key differences in catalytic residues were recognized in ARTD10 (Kleine et al. 2008). The catalytic motif His-Tyr-Glu is altered to His-Tyr-Ile in ARTD10, resulting in the ability to transfer only a single ADP-ribose to a substrate (Fig. 1). Mutating the glutamate of the catalytic motif in ARTD1 prevents the formation of polymers, supporting the essential role of this residue for poly-ADP-ribosylation activity, whereas ARTD10 does not acquire the capacity to form polymers when the glutamate is restored (Kleine et al. 2008; Marsischky et al. 1995). The catalytic glutamate in ARTD1 stabilizes the oxacarbenium ion transition state of NAD+, whereas in the model of the reaction catalyzed by ARTD10, a glutamate of a substrate takes over this function. However, unlike the catalytic glutamate in ARTD1, the glutamate of the substrate is subsequently ADP-ribosylated and as a result this modified residue is no longer available for a second round of catalysis. Thus, this mechanism––referred to as substrate-assisted catalysis––limits the activity of ARTD10 and the other subfamily members, ARTD7-8, ARTD11-12 and ARTD14-17 to mono-ADP-ribosylation (Kleine et al. 2008). Based on this model and chemical analysis of the reaction products it was proposed that ARTD10 selectively modifies acidic amino acids. More recent evidence suggests that ARTD family members may also be capable of modifying lysine residues (Altmeyer et al. 2009; Messner et al. 2010). These findings initiated a more general discussion about the specificity of different ARTDs which still continues, and led us to reevaluate the acceptor amino acids of ARTD10. An ARTD10 mutant, in which all lysines were exchanged to arginines, is still efficiently trans-automodified, indicating that lysines are at least not the exclusive acceptor sites (Rosenthal et al. 2013). With the evidence presently available we would argue that ARTD10 modifies acid amino acids, as it is unclear how this enzyme would be able of modifying both acidic side chains and lysines in the same catalytic center.
3 The Structure of ARTD10
The family of ARTDs was defined based on homology of the catalytic domains of these enzymes (Ame et al. 2004; Otto et al. 2005). The different ARTDs possess a great variety of additional domains and motifs besides the catalytic domain, which is thought to reflect the broad range of functions they carry out (Ame et al. 2004; Hottiger et al. 2010). Human ARTD10 contains 1025 amino acids, which approximate to an apparent molecular size of 150 kDa on SDS-PAGE. Less than 20% of the sequence contributes to the defining catalytic domain at the very C-terminus (Fig. 2a) (Yu et al. 2005), which is sufficient to mono-ADP-ribosylate ARTD10 substrates in vitro, comparable to the full-length protein (Kleine et al. 2008; Yu et al. 2005). ARTD10 furthermore contains several other domains and motifs, including an RNA recognition motif (RRM), two functional ubiquitin interaction motifs (UIM), sequences capable to promote nuclear targeting as well as nuclear export, and a small motif that mediates interaction with PCNA (Fig. 2a) (Kleine et al. 2008, 2012; Nicolae et al. 2014; Verheugd et al. 2013; Yu et al. 2005). It is conceivable that some of these domains orient ARTD10-mediated mono-ADP-ribosylation toward specific substrates in different compartments, guided for instance by the interaction with poly-ubiquitin (Herzog et al. 2013; Verheugd et al. 2013). ARTD10 hence seems suited to participate in a wide set of processes in cells.
Domains and structural motifs of ARTD10. a. The following domains and motifs are indicated: an RNA recognition motif (amino acids 11–85), a glycine-rich region (281–399), a caspase-6 cleavage site (C-terminal of Asp406), a nuclear localization region (435–528), a glutamate-rich region (588–697), a nuclear export signal (598–607), two ubiquitin interaction motifs (650–667 and 673–690), and a catalytic domain (818–1013), which contains a PCNA-interaction peptide motif (PIP box, amino acids 830–837). b. The known posttranslational modifications are indicated: Thr101 is phosphorylated by Cyclin E/CDK2; caspase-6 cleaves ARTD10 C-terminal of Asp406 during apoptosis; the catalytic domain is the major automodification region with Glu882 being one of the mono-ADP-ribosylation sites; Lys916 is both acetylated and ubiquitinated
4 Regulation of ARTD10
At present, little is known about the regulation of ARTD10. Several PTMs can be found on ARTD10, suggesting that multiple signaling pathways control it. The transferase domain of ARTD10 is mono-ADP-ribosylated by intermolecular automodification, presumably on multiple sites (Kleine et al. 2008; Yu et al. 2005). The only modification site that has been mapped to date is a glutamate (Glu882) (Fig. 2b). The mutation of Glu882 has no effect on catalytic activity and only a partial effect on automodification, in agreement with multiple ADP-ribosylation sites in ARTD10 (Kleine et al. 2008). Furthermore, two independent mass spectrometry approaches have suggested that ubiquitination of ARTD10 occurs at Lys916 in cells (Fig. 2b) (Hornbeck et al. 2012; Wagner et al. 2011). This represents a PTM, which might affect catalytic activity, as an ARTD10-K916I mutant yields a strongly reduced automodification signal (Yu et al. 2005). Intriguingly, another study found the same residue to be acetylated in cells (Choudhary et al. 2009). Competing ubiquitination and acetylation have been previously recognized as rivaling modulators of protein function and half-life (Caron et al. 2005; Li et al. 2012; Vervoorts et al. 2003; Yang and Seto 2008). ARTD10 activity could thus be regulated by ubiquitin ligases and acetyltransferases. Interestingly, K916 has only been introduced during primate evolution, while a highly conserved glutamine is present at the same site in lower species, including mice (Kim and Hahn 2012). Glutamines are commonly accepted as imperfect mimics of acetylated lysines (Fujimoto et al. 2012; Kamieniarz and Schneider 2009). Hence acetylation of K916 may preserve while ubiquitination of this residue may interfere with ARTD10 catalytic activity.
ARTD10 is also phosphorylated at multiple sites (our own unpublished findings), thus providing potential opportunities for regulation. In one report, Thr101 was identified as a phosphorylation site of cyclin-dependent kinases (Chou et al. 2006). The work performed in vitro suggested that phosphorylation at Thr101 enhances ARTD10 catalytic activity. We have also identified this phosphorylation site, but were unable to measure phosphorylation-dependent effects on catalytic activity (Fig. 2b). In summary the available evidence indicates that various enzymes modify ARTD10. Future work will have to address the pathways that potentially converge onto ARTD10 and to define the molecular consequences of different PTMs on the functions of ARTD10.
5 ARTD10 Shuttles Between the Cytoplasm and the Nucleus
ARTD10 substrates and interaction partners are situated both in the nucleus (e.g., c-MYC, RAN, and histones) and in the cytoplasm (e.g., GSK3β, NEMO/IKKγ, p62/SQSTM1, and PCNA) (Feijs et al. 2013b; Forst et al. 2013; Kleine et al. 2012; Nicolae et al. 2014; Verheugd et al. 2013; Yu et al. 2005). Under steady-state conditions in proliferating cells, little nuclear ARTD10 is detectable because of a strong nuclear export sequence (NES) that causes rapid export from the nucleus (Fig. 2a) (Kleine et al. 2012). Even when the NES is mutated or the CRM1-dependent export is inhibited by leptomycin B (Kudo et al. 1999), only an equal distribution is achieved rather than nuclear accumulation. Hence additional mechanisms must exist that retain ARTD10 in the cytoplasm (Kleine et al. 2012). ARTD10 could potentially be bound to a cytoplasmic protein or to a scaffold like poly-ubiquitin. Indeed, organization of ARTD10 into cytoplasmic bodies (Kleine et al. 2012; Vyas et al. 2013), associated with the autophagy adaptor p62 (Johansen and Lamark 2011; Nezis and Stenmark 2012) and ubiquitin, has been described (Kleine et al. 2012). The exchange of proteins in these bodies is swift and therefore does not seem to limit dynamic redistribution of ARTD10. ARTD10 might also be exported by a nuclear export pathway, which involves an exportin other than CRM1. Alternative routes have been established for proteins such as actin or 14-3-3σ (Mingot et al. 2004; Stuven et al. 2003). This would not change the dependence on a gradient of the nuclear GTP-binding protein RAN, as all known exportins form a ternary complex with their cargo and RAN-GTP in the nucleus, complexes that disintegrate in the cytoplasm after GTP hydrolysis (Guttler and Gorlich 2011). ARTD10 specifically mono-ADP-ribosylates the GTP-bound form of RAN, while RAN-GDP is not modified (Forst et al. 2013). Presently, it is not clear whether mono-ADP-ribosylation influences RAN function.
The nuclear uptake of ARTD10 relies on a conserved central region, which functions as a nuclear localization sequence (NLS) (Fig. 2a). However this domain does not contain a recognizable classical NLS and resembles no formerly described motif (Kleine et al. 2012). This ARTD10 domain can drive nuclear translocation when fused to EGFP-β-Gal, but unlike a classical NLS only a partial nuclear accumulation was observed. This suggests that this unconventional NLS may use a pathway that is not very efficient or that this domain has additional functions such as interacting with cytosolic factors and thus preventing efficient nuclear import. The intricacy of ARTD10 localization indicates that cells can regulate the subcellular distribution of ARTD10, although it is unclear how this is achieved. An interesting but purely speculative mechanism can be based on a study that finds ARTD10 mRNA specifically altered in blood cells by adenosine deaminases acting on RNA (ADARs) (He et al. 2011). These enzymes deaminate adenosines to inosines, which are recognized as guanosines by the translational machinery. In the case of ARTD10 this results in an amino acid substitution within the NLS region; the highly conserved Ser507 is exchanged by a glycine, which might affect the activity of the unconventional NLS of ARTD10. Together these findings demonstrate that ARTD10 is localized to both the nuclear and cytoplasmic compartments. Together with the known substrates and interaction partners highlighted above, it is highly likely that ARTD10 has functions in both compartments as described in more detail below. Moreover functions associated with the transport between nuclear and cytosolic compartments may also be conceivable for ARTD10.
6 Signaling Pathways Regulated by ARTD10
Different possibilities exist as to mono-ADP-ribosylation could influence target proteins; in analogy to other PTMs, effects on protein function and on interactions with other macromolecules are likely. Recent evidence supports both, i.e., ADP-ribose can function as allosteric regulator of enzyme activity and as docking site for distinct proteins (Feijs et al. 2013a, b; Forst et al. 2013). A reader domain of mono-ADP-ribosylated proteins was identified, indicating that mono-ADP-ribosylation promotes defined protein-protein interactions (Forst et al. 2013). This reader is a macrodomain, a highly conserved structure which is closely associated with ADP-ribosylation and ADP-ribose metabolism (for a more extensive summary of the currently known macrodomain-containing proteins and their functions see (Feijs et al. 2013a)). Two macrodomains in ARTD8 are capable to selectively interact with ARTD10-dependent mono-ADP-ribosylated substrates (Forst et al. 2013; Verheugd et al. 2013). It is noteworthy that ARTD8 functions also as mono-ADP-ribosyltransferase, which makes multistep mono-ADP-ribosylation events on proteins conceivable.
Apart from recognition of mono-ADP-ribosylation by specific protein domains, ARTD10-mediated modification of proteins can regulate substrate function directly, as exemplified by glycogen synthase kinase 3β (GSK3β). GSK3β is a well-investigated enzyme with established functions in WNT signaling, apoptosis, metabolism, neuronal development, immunity, and tumorigenesis (Beurel and Jope 2006; Beurel et al. 2010; Hur and Zhou 2010; Mills et al. 2011; Wu and Pan 2010). ARTD10 is able to mono-ADP-ribosylate GSK3β in vitro leading to a reduction of in vitro GSK3β kinase activity. This inhibition could not be overcome by increasing substrate concentration, implying that mono-ADP-ribosylation functions as an allosteric inhibitor of GSK3β.
Mono-ADP-ribosylation is currently not reliably measurable in cells due to a lack of suitable antibodies and mass spectrometry methods. To analyze GSK3β, its kinase activity was used as a surrogate parameter to evaluate ARTD10 activity in cells. Immunoprecipitated GSK3β showed reduced catalytic activity when co-expressed with ARTD10, while overexpression of the catalytically inactive mutant ARTD10-G888 W or knockdown of the endogenous ARTD10 significantly enhanced it, implying that modification of GSK3β indeed takes places in cells (Feijs et al. 2013b).
Recent findings indicate that mono-ADP-ribosylation is a reversible PTM (Jankevicius et al. 2013; Rosenthal et al. 2013; Sharifi et al. 2013). The enzymes capable of removing mono-ADP-ribose from protein substrates are MacroD1, MacroD2, and C6orf130/TARG1, which all contain a catalytically active macrodomain able to reverse ARTD10-dependent mono-ADP-ribosylation. Whether they can revert mono-ADP-ribosylation catalyzed by other mono-ARTDs needs to be determined. Moreover, MacroD2 was found to de-mono-ADP-ribosylate GSK3β, thereby restoring its catalytic activity (Rosenthal et al. 2013). This in vitro finding renders GSK3β modification by ARTD10 reversible, as would be expected from a mechanism that regulates signaling cascades. In cells, activation of GSK3β was also observed when MacroD2 was over-expressed, resulting together with the above summarized experiments in a strong case for endogenous mono-ADP-ribosylation of GSK3β and the functional relevance of this modification (Feijs et al. 2013b; Rosenthal et al. 2013). The precise understanding of the regulation of ARTD10-mediated mono-ADP-ribosylation and the antagonizing hydrolases will be important to identify the cellular contexts in which ARTD10’s activity is physiologically relevant.
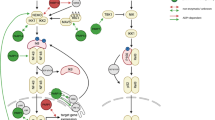
One pathway ARTD10 was postulated to interfere with is NF-κB signaling (Verheugd et al. 2013). The UIMs of ARTD10 were identified as interaction motifs for K63-linked poly-ubiquitin,, which is an important PTM within the NF-κB signal transduction pathway as well as other signaling networks (Grabbe et al. 2011). E3 ubiquitin ligases such as TRAF6 synthesize scaffolds of K63-linked poly-ubiquitin upon a number of stimuli, including inflammatory cytokines, which are essential to promote NF-κB translocation into the nucleus to enable transcription of its target genes (Fig. 3) (DiDonato et al. 2012; Oeckinghaus et al. 2011). ARTD10 functions as a repressor of an NF-κB reporter gene construct and chromatin embedded NF-κB target genes (Verheugd et al. 2013). This repression depends on the catalytic activity and the K63-linked poly-ubiquitin binding capability of ARTD10, as demonstrated by using the nonrepressive double mutant ARTD10-G888 W-ΔUIM, which lacks catalytic and K63-linked poly-ubiquitin binding activity. These findings suggest that in response to specific signals ARTD10 is recruited to K63-linked poly-ubiquitin scaffolds, resulting in mono-ADP-ribosylation of proteins in the vicinity of these scaffolds (Fig. 3). However, the abolishment of either ubiquitin binding or catalytic activity alone did not reduce ARTD10’s inhibitory potential towards NF-κB. One possible explanation for this observation might relate to the fact that the mutants were over-expressed. It is imaginable that abundant ARTD10-G888 W can compete for K63-linked poly-ubiquitin binding with factors essential for signal propagation, such as NEMO. Moreover, abundant ARTD10-ΔUIM may suffice to mono-ADP-ribosylate substrates in the pathway without the need of induced proximity. Both would result in the observed inhibitory effect. A role of ARTD10 in the NF-κB pathway was corroborated with ARTD10 knockdown experiments. These resulted in enhanced NF-κB signaling, suggesting that ARTD10 indeed downregulates this pathway in cells. Additionally, overexpression of ARTD10 leads to a specific decrease of the K63-linked poly-ubiquitin of NEMO (or IKKγ), an integral part of the IKK complex, which controls the NF-κB transcription factor (Fig. 3). NEMO serves as an adaptor of the complex to K63-linked poly-ubiquitin and is itself K63-poly-ubiquitinated, both features that are essential for NF-κB signal propagation (Chen 2012; Skaug et al. 2009). As NEMO is a substrate of ARTD10 in vitro and in cells (Verheugd et al. 2013), it will be interesting to see how its mono-ADP-ribosylation might interfere with its K63-linked poly-ubiquitination and how this relates to abortion of the signaling process.
ARTD10 regulates NF-κB signaling. A simplified version of the NF-κB signal transduction pathway is shown. In response to extracellular cues ubiquitin E3 ligases are activated that synthesize K63-linked poly-ubiquitin. These serve as scaffold for kinase complexes, including the TAK1 and the IKK complexes. The local vicinity allows TAK1 to phosphorylate and activate IKKβ. This kinase then modifies IκB, which results in its degradation and the subsequent activation of NF-κB transcription factors (left scheme). ARTD10 interferes with the activation of NF-κB by interacting with K63-linked poly-ubiquitin and by mono-ADP-ribosylating NEMO. This prevents poly-ubiquitination of NEMO, while enhancing it on the E3 ligase TRAF6 and on TAK1
Participation in signaling events so far is the area with the most substantial evidence for an involvement of ARTD10. Direct alteration of kinase activity as demonstrated for GSK3β indicates an intervention of ARTD10 in intracellular communication by phosphorylation, while the results from studying the NF-κB pathway suggest that ARTD10 can both read as well as interfere with K63-poly-ubiquitination of proteins. Together these results suggest that mono-ADP-ribosylation participates in crosstalk with other PTMs, including ubiquitination and phosphorylation.
7 A Role for ARTD10 in Transcription?
Different ARTDs such as ARTD1, ARTD8, and ARTD14 are involved in transcriptional regulation (Feijs et al. 2013c; Kraus 2008). There are only initial hints for a role of ARTD10 in transcription. ARTD10 was discovered in a search for binding partners of the oncogenic transcription factor MYC (Yu et al. 2005). Bimolecular fluorescence complementation later revealed that the MYC-ARTD10 interaction takes place in the nucleus (Kleine et al. 2012), but it has not been resolved whether the interaction occurs on DNA. Currently, the known functional relevance of the ARTD10 interaction with MYC is limited to co-transformation assays of rat embryo fibroblasts, in which ARTD10 acts as a suppressor of the transforming activity of MYC with HA-RAS (Yu et al. 2005). However, MYC is not a substrate of ARTD10, leaving it currently unknown whether ARTD10 controls MYC directly. Possible nuclear ARTD10 substrates that might influence transcription, such as histones, can be mono-ADP-ribosylated in vitro but it remains to be determined whether ARTD10 also modifies these proteins in cells (Yu et al. 2005). It is worth noting that there is substantial evidence that core histones are both mono- and poly-ADP-ribosylated in cells (Hassa et al. 2006). Nevertheless, a precise role of ARTD10 in transcription remains undefined at present.
8 ARTD10 Stimulates Apoptosis
HeLa cells with inducible expression of ARTD10 or the catalytically inactive point mutant ARTD10-G888 W have been used to characterize the influence of mono-ADP-ribosylation by ARTD10 on cell proliferation (Herzog et al. 2013). In these cells ARTD10 reduces proliferation and induces apoptosis, dependent on its catalytic activity. Additionally ARTD10 knockdown in U2OS cells leads to decreased apoptosis in response to DNA damaging treatments. Together these findings provide evidence for a pro-apoptotic function of endogenous ARTD10. Moreover, early during apoptosis ARTD10 itself is processed by caspase-6 (Fig. 2b), which reduces its pro-apoptotic potential (Herzog et al. 2013). Expression of the corresponding cleavage products did not result in cell death, even though the C-terminal fragment remained catalytically active. Further experiments revealed that indeed a physical connection between the transferase domain of ARTD10 and the N-terminal RRM is needed to stimulate any significant degree of apoptosis, although the exact mechanism remains to be clarified.
What might be the role of the RRM for ARTD10-induced apoptosis? RRMs function in many different processes that regulate and are dependent on RNA and have also been described more recently to mediate protein-protein interactions (Maris et al. 2005). A potential answer to why ARTD10’s RRM might be important could therefore be a dependence of ARTD10 on its RRM to interact with certain substrates, either directly or mediated by RNA. Truncation of the RRM also results in a loss of the formation of p62-associated ARTD10 bodies in the cytoplasm (Kleine et al. 2012). These structures are not identical with well-studied RNA-containing particles like stress granules or P-bodies. The p62 bodies, which are also positive for ubiquitin, have been implicated as signaling hubs that regulate apoptosis and autophagy (Johansen and Lamark 2011; Kleine et al. 2012; Nezis and Stenmark 2012).
Why is ARTD10, which promotes apoptosis, a target of caspases? Once apoptosis is proceeding, protein biosynthesis is rapidly down-regulated (Clemens et al. 2000). Accordingly, RNA binding and processing proteins such as splicing factors become major targets of caspases, resulting in impeding of mRNA maturation and transport to the cytoplasm (Thiede et al. 2001; Thomas and Lieberman 2013). Based on the findings that ARTD10 shuttles between nuclear and cytoplasmic compartments and the presence of an RRM, we speculate that ARTD10 might be part of a regulatory network that integrates the control of RNA processing with programmed cell death.
If ARTD10 stimulates apoptosis through mono-ADP-ribosylation, which substrates could contribute to such a function? Interestingly, the ARTD10 target GSK3β has been thoroughly investigated as an alternate regulator of apoptosis with the capability to stimulate the intrinsic pathway, while inhibiting the extrinsic pathway (Beurel and Jope 2006). GSK3β knockout mice die of massive hepatic apoptosis during development, while GSK3α−/− mice are vital (Gomez-Sintes et al. 2011; Hoeflich et al. 2000). It has been suggested that the GSK3β-dependent activation of the NF-κB survival response is at least in part responsible for the observed effect (Hoeflich et al. 2000). As ARTD10 negatively regulates GSK3β by mono-ADP-ribosylation and represses NF-κB signaling by targeting NEMO, the inhibition of both GSK3β and NEMO might link ARTD10 to apoptosis (Feijs et al. 2013b; Verheugd et al. 2013). Additional ARTD10 interaction partners and substrates might contribute to the pro-apoptotic phenotype. The nuclear transport protein RAN, which is an ARTD10 substrate (Forst et al. 2013), has been suggested to prevent the nuclear import of NF-κB early during apoptosis to enhance apoptotic progression (Wong et al. 2009). Moreover, cytosolic p62-ubiquitin bodies are implicated as signal organizing centers during apoptosis. They can either recruit TRAF6 to subsequently activate NF-κB by auto-K63-poly-ubiquitination or aggregate ubiquitinated caspase-8, which promotes its activation and subsequent stimulation of apoptosis (Moscat and Diaz-Meco 2009), suggesting that the ARTD10 interaction partner p62 participates in controlling cell survival. Together these findings offer additional entry points for ARTD10 in the control of apoptosis that are in need of evaluation.
9 DNA Repair
One of the key activities of ARTD1 is its function in DNA repair processes. ARTD1 recognizes damaged DNA through its Zn-fingers, resulting in the allosteric activation of its catalytic domain. Subsequently, ADP-ribose polymers are synthesized on ARTD1 itself as well as other proteins, including core histones (De Vos et al. 2012). These polymers act as scaffolds to recruit DNA repair proteins through ADP-ribose polymer binding domains (Kleine and Luscher 2009). ARTD2/PARP2 and ARTD3/PARP3 are two additional family members that are involved in DNA repair processes (De Vos et al. 2012). Thus poly-ADP-ribosylation plays a major role in different DNA repair pathways. Little is known about the role of mono-ADP-ribosylation in DNA repair. A very recent study identified PCNA, which is associated with a large number of processes associated with genomic stability (Groth et al. 2007; Moldovan et al. 2007), as an interaction partner of ARTD10. It possesses a PCNA-interacting peptide (PIP) box near the N-terminal end of the catalytic domain (Fig. 2a) (Nicolae et al. 2014). The PIP box is known to interact with a number of proteins and thus is important to control and integrate PCNA activities (Moldovan et al. 2007). Indeed ARTD10 interacts through its PIP box with PCNA, an interaction that seems to be required to promote genomic stability and tolerance to DNA damage (Nicolae et al. 2014). ARTD10 is also important for PCNA-dependent translesion DNA synthesis and associated mutagenesis, requiring an active catalytic domain. The interaction of ARTD10 with PCNA is further promoted by the UIMs, which enhance the interaction with ubiquitinated PCNA. Together these findings support the notion that in addition to poly-ADP-ribosylation also mono-ADP-ribosylation participates in DNA repair processes. It will be of interest to determine whether and if how these two types of ADP-ribosylation reactions collaborate.
10 Clues Toward Additional Biological Functions of ARTD10
Presently our understanding of ARTD10 resembles a patchwork of promising suggestions yet little substantial evidence. To further evaluate potential functions of ARTD10, it is worth to consider the information that is available connecting ARTD10 with distinct cellular and organismal processes. In the following, we summarize the preliminary indications of interaction of ARTD10 with the immune system, metabolism, and with cancer.
10.1 ARTD10 Levels Differ Between Tissues
In the initial report on ARTD10, tissue-specific expression was analyzed by northern blotting of human samples (Yu et al. 2005). ARTD10 mRNA was detected in all tested tissue samples with increased expression in thymus and spleen when compared to actin mRNA. This indicated enhanced expression of ARTD10 in cells of the immune system. Indeed ARTD10 protein levels are elevated in cell lines originating from the hematopoietic system (Yu et al. 2005). Information from publicly available human expression datasets largely confirms these results (Fig. 4). Also array data from mice and rats are available, which include a broader set of tissue samples (Fig. 4). Besides expression in thymus and spleen, high levels of ARTD10 are found in other tissues in these studies, including adipose tissue, the Langerhans islets of the pancreas, the adrenal glands, the small intestine, the liver, and the kidneys. Low expression of ARTD10 in the brain was measured in multiple studies, with the pronounced exception of the pituitary gland (Hindmarch et al. 2006; Hovatta et al. 2007), a tissue that shows among the highest expression levels. Importantly this is not limited to the adenohypophysis, which is not of neuronal origin, but was also measured in the neurohypophysis (Hindmarch et al. 2006). Thus ARTD10 expression is predicted to be widespread (Fig. 4).
Relative ARTD10 expression in different tissues. The heat map visualizes mRNA expression data from eight different studies, including one study focused on human ARTD10 (1, Yu et al. 2005), three genome-wide human expression data sets (2, {Yanai, 2005 #912}, http://www.ebi.ac.uk/gxa/, E-GEOD-803; 3, {Liu, 2008 #913}; 4, {Krupp, 2012 #914}), RNA expression data from the Human Protein Atlas (5, http://www.proteinatlas.org), and three genome-wide mouse expression data sets (6, {Su, 2002 #915}; 7, {Thorrez, 2008 #916}, http://www.ebi.ac.uk/gxa/, E-GEOD-9954; 8, {Thorrez, 2011 #917}, http://www.ebi.ac.uk/gxa/, E-GEOD-24940). ARTD10 expression in a given tissue was judged as not detectable, low, intermediate, or high compared to ARTD10 expression in other tissues within a given study. In the top row these results are combined to give a general impression in which organs ARTD10 is expressed preferentially. Tissues that were not studied are indicated as not applicable (n/a)
10.2 ARTD10 is Upregulated in Response to Inflammatory Stimuli
ARTD10, together with certain other ARTDs, belongs to a group of genes, which are upregulated in sooty mangabeys and rhesus macaques in response to infection with simian immunodeficiency virus (Bosinger et al. 2009). The enhanced expression was measured on day 7 and 10 after infection, which coincided with a general activation of interferon (IFN) response genes as well as pro-apoptotic and cell cycle regulating genes. In other studies, the response of monocytes to phagocytosed live borrelia burgdorferi was investigated compared to toll-like receptor activation by borrelia lysates. Two studies demonstrated a dose-dependent induction of apoptosis upon borrelia phagocytosis, but not by borrelia lysates, in accordance with activation of IFN production and upregulation of a gene set including ARTD10 (Cruz et al. 2008; Salazar et al. 2009). Furthermore, a noncytopathic variant of the Venezuelan equine encephalitis virus (an alphavirus), which is cleared from wild-type mouse embryo fibroblasts (MEFs) but not from MEFs lacking IFNα/β receptors, appears to regulate ARTD10 expression (Atasheva et al. 2012). ARTD10 and ARTD12 expression are up-regulated upon infection with the virus or IFNβ treatment after 24 h, whereas no effect was observed after infection in type I IFN receptor negative MEFs, suggesting that the stimulation of the IFN response triggered ARTD10 expression. Moreover, ARTD10 as well as ARTD family members appear to function as strong inhibitors of protein biosynthesis and inhibit alphavirus replication (Atasheva et al. 2014). ARTD10 is also one of 46 genes, which display high expression after IFNα treatment of a liver cell line (Mahmoud et al. 2011). In support an ARTD10 promoter-reporter gene construct is readily responsive to infection with the Newcastle disease virus, a strong inducer of type I IFNs. ChIP-seq analysis for IRF1 binding sites, a downstream effector of IFNα, revealed that this transcription factor binds to the ARTD10 promoter (Shi et al. 2011). Finally, one study presented evidence for a downregulation of ARTD10 upon ectopic expression of the influenza protein NS1 (Yu et al. 2011), which may be an indirect effect as NS1 is known to inhibit type I IFN production (Hale et al. 2008). Although these studies rely largely on the analysis of RNA, together these findings strongly implicate ARTD10 in anti-viral response.
10.3 ARTD10 Might be Involved in Lipid Metabolism
ARTD10 has been identified as a locus associated with lipid traits in a large meta-analysis summarizing 46 genome-wide association studies (Teslovich et al. 2010). Using a sample of >100,000 individuals of European ancestry 95 loci were identified to be significantly associated with plasma lipids, with 22 being associated with changes in LDL cholesterol levels in the blood. Among these, the lead single nucleotide polymorphism (SNP) rs11136341 is associated with increased LDL and total cholesterol. This SNP maps to the beginning of the Plectin1 gene and is roughly 5000 base pairs downstream of the ARTD10 gene, consistent with the known overlap of the two genes (Fuchs et al. 2009; Lesniewicz et al. 2005). The potential relevance of ARTD10 was addressed using knockdown studies, which revealed that this resulted in a substantial decrease in apolipoprotein B (APOB) secretion of liver cells (Shen et al. 2012). The APOB gene was also described as one of 22 loci associated with a lead SNP of increased LDL levels (Teslovich et al. 2010). APOB is the primary lipoprotein of chylomicrons and LDL particles, its overexpression is associated with high LDL and low HDL blood levels and it is regarded as a risk factor of atherosclerosis (Benn 2009; McCormick et al. 1996). Here, it may also be of note that macrophages treated with oxidized LDL show a significant downregulation of ARTD10 expression determined by mass spectrometry (Kang et al. 2009). Further experimentation will be required to firmly establish ARTD10 in the control of lipid metabolism.
10.4 ARTD10 in Tumor Biology?
Could ARTD10 also be relevant for tumor cells, similar to other ARTDs that have been implicated in tumorigenesis (Feijs et al. 2013c; Scarpa et al. 2013)? Some observations are in support of such a hypothesis. ARTD10 expression varies considerably in different tumor cell lines and its overexpression inhibits transformation of rat embryo fibroblasts (Yu et al. 2005). The ARTD10 gene is also located in the same part of chromosome 8 as its oncogenic interaction partner MYC (8q24), which could result in simultaneous deregulation by chromosomal aberrations. Thus, far ARTD10 expression in tumors has not been analyzed systematically. However one study exists, which noticed a tenfold ARTD10 downregulation in a set of 86 samples of gastric cancer compared to normal gastric tissue (Wong et al. 2009). As ARTD10 displays anti-proliferative and pro-apoptotic properties at least in some tumor cell lines, it may function as a tumor suppressor, a hypothesis that is in need of verification.
11 Conclusions and Outlook
The expression data summarized here provide indications about the cell types and tissues in which ARTD10 may be relevant. The preferential expression of ARTD10 in the thymus and in the spleen together with its induction upon inflammatory or immunogenic stimuli suggests a role in innate immunity (Fig. 5), an area where other ARTDs have been established (Welsby et al. 2012). Expression in adipose tissue, liver, and pancreas together with data that link ARTD10 to lipoproteins may point to a role in metabolism (Fig. 5). Studies addressing roles of ARTD10 in NF-κB signaling, apoptosis, and DNA damage also indicate that this protein fulfills multiple tasks (Fig. 5), perhaps regulated by extracellular cues such as IFNα. The mechanisms through which ARTD10 functions remain largely unclear but might become better understood once for example more target proteins are identified. However, the recent identification of mono-ADP-ribosylhydrolases indicates that mono-ADP-ribosylation is a reversible PTM and the direct regulation of GSK3β underlines that it can have profound effects on the activities of substrate proteins.
Potential involvement of ARTD10 in cellular and organismal processes. Biochemical studies have implicated ARTD10 in different aspects of cell physiology such as apoptosis, signaling, transcription and DNA repair. At the organismal level, these ARTD10-mediated cellular functions may be the underlying mechanisms to modulate processes such as metabolism, innate immunity, and cancer (for more discussion see the main text)
An effective way to expand and deepen our knowledge of mono-ADP-ribosylation in general and mono-ADP-ribosylation by ARTD10 in particular will be the development of more elaborate tools to study mono-ADP-ribosylation in cells. For example the development of antibodies that specifically recognize mono-ADP-ribosylated proteins would allow analyzing the distribution, the dynamics, and the consequences of mono-ADP-ribosylation. Immediate applications of such tools would be to address the regulation of ARTD10, defining in vivo substrates and ultimately to understand the functional consequences of this PTM.
Although we have begun to understand basic characteristics of ARTD10 and recent findings have shed light on some of its biological properties, many questions remain open. We imagine that the studies addressing these will provide novel insight into the role of ARTD10 and mono-ADP-ribosylation in different fundamental processes, including inflammation and immunity and tumorigenesis. As ARTD1/PARP1 inhibitors are successfully entering clinical application (Rouleau et al. 2010), it is likely that other ARTD enzymes, including ARTD10, will serve as targets for therapeutic applications. Studying these enzymes thus provides opportunities to expand our knowledge on basic physiological processes and to translate this information into improving human health.
Abbreviations
- ADAR:
-
Adenosine deaminases acting on RNA
- APOB:
-
Apolipoprotein B
- ARTC:
-
ADP-ribosyltransferases Cholera toxin-like
- ARTD:
-
ADP-ribosyltransferase Diphtheria toxin-like
- GSK3β:
-
Glycogen synthase kinase 3β
- IFN:
-
Interferon
- MEFs:
-
Mouse embryo fibroblasts
- NES:
-
Nuclear export sequence
- NLS:
-
Nuclear localization sequence
- PIP:
-
PCNA-interacting peptide
- PTM:
-
Posttranslational modification
- RRM:
-
RNA recognition motif
- SNP:
-
Single nucleotide polymorphism
- UIM:
-
Ubiquitin interaction motif
References
Altmeyer M, Messner S, Hassa PO et al (2009) Molecular mechanism of poly(ADP-ribosyl)ation by PARP1 and identification of lysine residues as ADP-ribose acceptor sites. Nucleic acids research 37:3723–3738
Ame JC, Spenlehauer C, de Murcia G (2004) The PARP superfamily. Bioessays 26:882–893
Atasheva S, Akhrymuk M, Frolova EI et al (2012) New PARP gene with an anti-alphavirus function. J Virol 86:8147–8160
Atasheva S, Frolova EI, Frolov I (2014) Interferon-stimulated poly(ADP-Ribose) polymerases are potent inhibitors of cellular translation and virus replication. J Virol 88:2116–2130
Barkauskaite E, Jankevicius G, Ladurner AG et al (2013) The recognition and removal of cellular poly(ADP-ribose) signals. FEBS J 280:3491–3507
Benn M (2009) Apolipoprotein B levels, APOB alleles, and risk of ischemic cardiovascular disease in the general population, a review. Atherosclerosis 206:17–30
Beurel E, Jope RS (2006) The paradoxical pro- and anti-apoptotic actions of GSK3 in the intrinsic and extrinsic apoptosis signaling pathways. Prog Neurobiol 79:173–189
Beurel E, Michalek SM, Jope RS (2010) Innate and adaptive immune responses regulated by glycogen synthase kinase-3 (GSK3). Trends in immunology 31:24–31
Bosinger SE, Li Q, Gordon SN et al (2009) Global genomic analysis reveals rapid control of a robust innate response in SIV-infected sooty mangabeys. The Journal of clinical investigation 119:3556–3572
Caron C, Boyault C, Khochbin S (2005) Regulatory cross-talk between lysine acetylation and ubiquitination: role in the control of protein stability. Bioessays 27:408–415
Chen ZJ (2012) Ubiquitination in signaling to and activation of IKK. Immunological reviews 246:95–106
Chou HY, Chou HT, Lee SC (2006) CDK-dependent activation of poly(ADP-ribose) polymerase member 10 (PARP10). J Biol Chem 281:15201–15207
Choudhary C, Kumar C, Gnad F et al (2009) Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science 325:834–840
Clemens MJ, Bushell M, Jeffrey IW et al (2000) Translation initiation factor modifications and the regulation of protein synthesis in apoptotic cells. Cell Death Differ 7:603–615
Cruz AR, Moore MW, La Vake CJ et al (2008) Phagocytosis of Borrelia burgdorferi, the Lyme disease spirochete, potentiates innate immune activation and induces apoptosis in human monocytes. Infect Immun 76:56–70
D’Amours D, Desnoyers S, D’Silva I et al (1999) Poly(ADP-ribosyl)ation reactions in the regulation of nuclear functions. Biochem J 342 (Pt 2):249–268
De Vos M, Schreiber V, Dantzer F (2012) The diverse roles and clinical relevance of PARPs in DNA damage repair: current state of the art. Biochem Pharmacol 84:137–146
Deng Q, Barbieri JT (2008) Molecular mechanisms of the cytotoxicity of ADP-ribosylating toxins. Annu Rev Microbiol 62:271–288
Di Girolamo M, Dani N, Stilla A et al (2005) Physiological relevance of the endogenous mono(ADP-ribosyl)ation of cellular proteins. Febs J 272:4565–4575
DiDonato JA, Mercurio F, Karin M (2012) NF-kappaB and the link between inflammation and cancer. Immunol Rev 246:379–400
Feijs KL, Forst AH, Verheugd P et al (2013a) Macrodomain-containing proteins: regulating new intracellular functions of mono(ADP-ribosyl)ation. Nat Rev Mol Cell Biol 14:443–451
Feijs KL, Kleine H, Braczynski A et al (2013b) ARTD10 substrate identification on protein microarrays: regulation of GSK3beta by mono-ADP-ribosylation. Cell Commun Signal 11:5
Feijs KL, Verheugd P, Luscher B (2013c) Expanding functions of intracellular resident mono-ADP-ribosylation in cell physiology. FEBS J 280:3519–3529
Forst AH, Karlberg T, Herzog N et al (2013) Recognition of mono-ADP-ribosylated ARTD10 substrates by ARTD8 macrodomains. Structure 21:462–475
Fuchs P, Zorer M, Reipert S et al (2009) Targeted inactivation of a developmentally regulated neural plectin isoform (plectin 1c) in mice leads to reduced motor nerve conduction velocity. J Biol Chem 284:26502–26509
Fujimoto H, Higuchi M, Koike M et al (2012) A possible overestimation of the effect of acetylation on lysine residues in KQ mutant analysis. J Comput Chem 33:239–246
Gibson BA, Kraus WL (2012) New insights into the molecular and cellular functions of poly(ADP-ribose) and PARPs. Nat Rev Mol Cell Biol 13:411–424
Gomez-Sintes R, Hernandez F, Lucas JJ et al (2011) GSK-3 Mouse Models to Study Neuronal Apoptosis and Neurodegeneration. Front Mol Neurosci 4:45
Grabbe C, Husnjak K, Dikic I (2011) The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol 12:295–307
Groth A, Rocha W, Verreault A et al (2007) Chromatin challenges during DNA replication and repair. Cell 128:721–733
Guttler T, Gorlich D (2011) Ran-dependent nuclear export mediators: a structural perspective. EMBO J 30:3457–3474
Hale BG, Randall RE, Ortin J et al (2008) The multifunctional NS1 protein of influenza A viruses. J Gen Virol 89:2359–2376
Hassa PO, Haenni SS, Elser M et al (2006) Nuclear ADP-ribosylation reactions in mammalian cells: where are we today and where are we going? Microbiol Mol Biol Rev 70:789–829
He T, Wang Q, Feng G et al (2011) Computational detection and functional analysis of human tissue-specific A-to-I RNA editing. PLoS ONE 6:e18129
Herzog N, Hartkamp JD, Verheugd P et al (2013) Caspase-dependent cleavage of the mono-ADP-ribosyltransferase ARTD10 interferes with its pro-apoptotic function. FEBS J 280:1330–1343
Hindmarch C, Yao S, Beighton G et al (2006) A comprehensive description of the transcriptome of the hypothalamoneurohypophyseal system in euhydrated and dehydrated rats. Proc Natl Acad Sci U S A 103:1609–1614
Hoeflich KP, Luo J, Rubie EA et al (2000) Requirement for glycogen synthase kinase-3beta in cell survival and NF-kappaB activation. Nature 406:86–90
Hornbeck PV, Kornhauser JM, Tkachev S et al (2012) PhosphoSitePlus: a comprehensive resource for investigating the structure and function of experimentally determined post-translational modifications in man and mouse. Nucleic Acids Res 40:D261–D270
Hottiger MO, Hassa PO, Luscher B et al (2010) Toward a unified nomenclature for mammalian ADP-ribosyltransferases. Trends Biochem Sci 35:208–219
Hovatta I, Zapala MA, Broide RS et al (2007) DNA variation and brain region-specific expression profiles exhibit different relationships between inbred mouse strains: implications for eQTL mapping studies. Genome Biol 8:R25
Hur EM, Zhou FQ (2010) GSK3 signalling in neural development. Nat Rev Neurosci 11:539–551
Jankevicius G, Hassler M, Golia B et al (2013) A family of macrodomain proteins reverses cellular mono-ADP-ribosylation. Nature structural & molecular biology 20:508–514
Johansen T, Lamark T (2011) Selective autophagy mediated by autophagic adapter proteins. Autophagy 7:279–296
Kamieniarz K, Schneider R (2009) Tools to tackle protein acetylation. Chem Biol 16:1027–1029
Kang JH, Ryu HS, Kim HT et al (2009) Proteomic analysis of human macrophages exposed to hypochlorite-oxidized low-density lipoprotein. Biochim Biophys Acta 1794:446–458
Kim DS, Hahn Y (2012) Gains of ubiquitylation sites in highly conserved proteins in the human lineage. BMC Bioinformatics 13:306
Kleine H, Herrmann A, Lamark T et al (2012) Dynamic subcellular localization of the mono-ADP-ribosyltransferase ARTD10 and interaction with the ubiquitin receptor p62. Cell Commun Signal 10:28
Kleine H, Luscher B (2009) Learning how to read ADP-ribosylation. Cell 139:17–19
Kleine H, Poreba E, Lesniewicz K et al (2008) Substrate-assisted catalysis by PARP10 limits its activity to mono-ADP-ribosylation. Mol Cell 32:57–69
Kraus WL (2008) Transcriptional control by PARP-1: chromatin modulation, enhancer-binding, coregulation, and insulation. Curr Opin Cell Biol 20:294–302
Krishnakumar R, Kraus WL (2010) The PARP side of the nucleus: molecular actions, physiological outcomes, and clinical targets. Mol Cell 39:8–24
Kudo N, Matsumori N, Taoka H et al (1999) Leptomycin B inactivates CRM1/exportin 1 by covalent modification at a cysteine residue in the central conserved region. Proc Natl Acad Sci U S A 96:9112–9117
Laing S, Unger M, Koch-Nolte F et al (2011) ADP-ribosylation of arginine. Amino Acids 41:257–269
Lesniewicz K, Luscher-Firzlaff J, Poreba E et al (2005) Overlap of the gene encoding the novel poly(ADP-ribose) polymerase Parp10 with the plectin 1 gene and common use of exon sequences. Genomics 86:38–46
Li H, Wittwer T, Weber A et al (2012) Regulation of NF-kappaB activity by competition between RelA acetylation and ubiquitination. Oncogene 31:611–623
Luo X, Kraus WL (2012) On PAR with PARP: cellular stress signaling through poly(ADP-ribose) and PARP-1. Genes Dev 26:417–432
Mahmoud L, Al-Saif M, Amer HM et al (2011) Green fluorescent protein reporter system with transcriptional sequence heterogeneity for monitoring the interferon response. Journal of virology 85:9268–9275
Maris C, Dominguez C, Allain FH (2005) The RNA recognition motif, a plastic RNA-binding platform to regulate post-transcriptional gene expression. The FEBS journal 272:2118–2131
Marsischky GT, Wilson BA, Collier RJ (1995) Role of glutamic acid 988 of human poly-ADP-ribose polymerase in polymer formation. Evidence for active site similarities to the ADP-ribosylating toxins. J Biol Chem 270:3247–3254
McCormick SP, Ng JK, Veniant M et al (1996) Transgenic mice that overexpress mouse apolipoprotein B. Evidence that the DNA sequences controlling intestinal expression of the apolipoprotein B gene are distant from the structural gene. J Biol Chem 271:11963–11970
Messner S, Altmeyer M, Zhao H et al (2010) PARP1 ADP-ribosylates lysine residues of the core histone tails. Nucleic Acids Res 38:6350–6362
Messner S, Hottiger MO (2011) Histone ADP-ribosylation in DNA repair, replication and transcription. Trends Cell Biol 21:534–542
Mills CN, Nowsheen S, Bonner JA et al (2011) Emerging roles of glycogen synthase kinase 3 in the treatment of brain tumors. Front Mol Neurosci 4:47
Mingot JM, Bohnsack MT, Jakle U et al (2004) Exportin 7 defines a novel general nuclear export pathway. EMBO J 23:3227–36
Moldovan GL, Pfander B, Jentsch S (2007) PCNA, the maestro of the replication fork. Cell 129:665–679
Moscat J, Diaz-Meco MT (2009) p62 at the crossroads of autophagy, apoptosis, and cancer. Cell 137:1001–1004
Nezis IP, Stenmark H (2012) p62 at the interface of autophagy, oxidative stress signaling, and cancer. Antioxid Redox Signal 17:786–793
Nicolae CM, Aho ER, Vlahos AH et al (2014) The ADP-ribosyltransferase PARP10/ARTD10 interacts with Proliferating Cell Nuclear Antigen (PCNA) and is required for DNA damage tolerance. J Biol Chem
Oeckinghaus A, Hayden MS, Ghosh S (2011) Crosstalk in NF-kappaB signaling pathways. Nat Immunol 12:695–708
Otto H, Reche PA, Bazan F et al (2005) In silico characterization of the family of PARP-like poly(ADP-ribosyl)transferases (pARTs). BMC Genomics 6:139
Rosenthal F, Feijs KL, Frugier E et al (2013) Macrodomain-containing proteins are new mono-ADP-ribosylhydrolases. Nat Struct Mol Biol 20:502–507
Rouleau M, Patel A, Hendzel MJ et al (2010) PARP inhibition: PARP1 and beyond. Nature reviews. Cancer 10:293–301
Salazar JC, Duhnam-Ems S, La Vake C et al (2009) Activation of human monocytes by live Borrelia burgdorferi generates TLR2-dependent and -independent responses which include induction of IFN-beta. PLoS pathogens 5:e1000444
Scarpa ES, Fabrizio G, Di Girolamo M (2013) A role of intracellular mono-ADP-ribosylation in cancer biology. FEBS J 280:3551–3562
Schreiber V, Dantzer F, Ame JC et al (2006) Poly(ADP-ribose): novel functions for an old molecule. Nat Rev Mol Cell Biol 7:517–528
Sharifi R, Morra R, Denise Appel C et al (2013) Deficiency of terminal ADP-ribose protein glycohydrolase TARG1/C6orf130 in neurodegenerative disease. The EMBO journal
Shen X, Wang W, Wang L et al (2012) Identification of genes affecting apolipoprotein B secretion following siRNA-mediated gene knockdown in primary human hepatocytes. Atherosclerosis 222:154–157
Shi L, Perin JC, Leipzig J et al (2011) Genome-wide analysis of interferon regulatory factor I binding in primary human monocytes. Gene 487:21–28
Skaug B, Jiang X, Chen ZJ (2009) The role of ubiquitin in NF-kappaB regulatory pathways. Annu Rev Biochem 78:769–796
Stuven T, Hartmann E, Gorlich D (2003) Exportin 6: a novel nuclear export receptor that is specific for profilin.actin complexes. EMBO J 22:5928–5940
Teslovich TM, Musunuru K, Smith AV et al (2010) Biological, clinical and population relevance of 95 loci for blood lipids. Nature 466:707–713
Thiede B, Dimmler C, Siejak F et al (2001) Predominant identification of RNA-binding proteins in Fas-induced apoptosis by proteome analysis. J Biol Chem 276:26044–26050
Thomas MP, Lieberman J (2013) Live or let die: posttranscriptional gene regulation in cell stress and cell death. Immunol Rev 253:237–252
Verheugd P, Forst AH, Milke L et al (2013) Regulation of NF-kappaB signalling by the mono-ADP-ribosyltransferase ARTD10. Nature communications 4:1683
Vervoorts J, Luscher-Firzlaff JM, Rottmann S et al (2003) Stimulation of c-MYC transcriptional activity and acetylation by recruitment of the cofactor CBP. EMBO Rep 4:484–490
Vyas S, Chesarone-Cataldo M, Todorova T et al (2013) A systematic analysis of the PARP protein family identifies new functions critical for cell physiology. Nature communications 4:2240
Wagner SA, Beli P, Weinert BT et al (2011) A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles. Mol Cell Proteomics 10:M111 013284
Welsby I, Hutin D, Leo O (2012) Complex roles of members of the ADP-ribosyl transferase super family in immune defences: looking beyond PARP1. Biochem Pharmacol 84:11–20
Wong CH, Chan H, Ho CY et al (2009) Apoptotic histone modification inhibits nuclear transport by regulating RCC1. Nat Cell Biol 11:36–45
Wu D, Pan W (2010) GSK3: a multifaceted kinase in Wnt signaling. Trends in biochemical sciences 35:161–168
Yang XJ, Seto E (2008) Lysine acetylation: codified crosstalk with other posttranslational modifications. Mol Cell 31:449–461
Yu M, Schreek S, Cerni C et al (2005) PARP-10, a novel Myc-interacting protein with poly(ADP-ribose) polymerase activity, inhibits transformation. Oncogene 24:1982–1993
Yu M, Zhang C, Yang Y et al (2011) The interaction between the PARP10 protein and the NS1 protein of H5N1 AIV and its effect on virus replication. Virology journal 8:546
Acknowledgments
We thank Andrew Jefferson for editing the manuscript. The work in our laboratory was supported by a Mildred Scheel Stipend of the German Cancer Aid (to MK), the START program of the Medical School of the RWTH Aachen University, and by the Deutsche Forschungsgemeinschaft DFG (to BL).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer International Publishing Switzerland
About this chapter
Cite this chapter
Kaufmann, M., Feijs, K.L.H., Lüscher, B. (2014). Function and Regulation of the Mono-ADP-Ribosyltransferase ARTD10. In: Koch-Nolte, F. (eds) Endogenous ADP-Ribosylation. Current Topics in Microbiology and Immunology, vol 384. Springer, Cham. https://doi.org/10.1007/82_2014_379
Download citation
DOI: https://doi.org/10.1007/82_2014_379
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-10770-7
Online ISBN: 978-3-319-10771-4
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)