Abstract
Recent years have witnessed an increased appreciation of the extent and relevance of strain-to-strain variation in Mycobacterium tuberculosis. This paradigm shift can largely be attributed to an improved understanding of the global population structure of this organism, and to the realisation that the various members of the M. tuberculosis complex (MTBC) harbour more genetic diversity than previously realised. Moreover, many studies using experimental models of infection have demonstrated that MTBC diversity translates into significant differences in immunogenecity and virulence . However, linking these experimental phenotypes to relevant clinical phenotypes has been difficult, and to date, largely unsuccessful. Nevertheless, emerging high-throughput technologies, in particular next-generation sequencing , offer new opportunities, and have already lead to important new insights. Given the complexity of the host-pathogen interaction in tuberculosis, systems approaches will be key to define the role of MTBC diversity in the fight against one of humankind’s most important pathogens.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
These keywords were added by machine and not by the authors. This process is experimental and the keywords may be updated as the learning algorithm improves.
1 Introduction
The outcome of tuberculosis (TB) infection and disease is extraordinarily diverse, ranging from lifelong asymptomatic infection to pulmonary TB and disseminated disease. In the past, this variable outcome has largely been attributed to host and environmental factors, but there is mounting evidence suggesting variation in the causative agent might also play a role. Indeed, the fact that this book, which is primarily focusing on the pathogenesis and host-pathogen interaction in TB, includes a chapter on “genetic diversity in M. tuberculosis” is a testimony to the recent paradigm shift with respect to the importance of studying strain variation (Comas and Gagneux 2009). Early sequencing studies revealed little DNA diversity across clinical isolates (Musser et al. 2000; Sreevatsan et al. 1997) leading to the dogma that strain variation in M. tuberculosis was “negligible” and therefore clinically irrelevant. Even though genetic diversity in M. tuberculosis is limited compared to other bacteria (Achtman 2008), this does not necessarily mean that the existing variability, as minor it may be, will not translate into any relevant phenotypes. Already in the 1960s, Mitchison and colleagues observed differences in virulence in clinical strains of M. tuberculosis when infecting guinea-pigs (Mitchison et al. 1960). At the time, no molecular tools were available to classify these strains into phylogenetically meaningful groupings. When the first genotyping tools became available in the early 1990s (van Embden et al. 1993), epidemiologist noticed very soon that M. tuberculosis strains also differed in their propensity to spread between individuals, and that some of these strains were associated with prolonged outbreaks (Rajakumar et al. 2004; Valway et al. 1998; Zhang et al. 1999). In some cases, experimental studies identified molecular features that might contribute to the success of outbreak strains (Newton et al. 2006; Reed et al. 2004). However, the relevance of these characteristics relative to non-bacterial factors remains unclear.
Over the last 10 years, several reviews have been published on the extent and relevance of strain diversity in M. tuberculosis (Gagneux and Small 2007; Kato-Maeda et al. 2001a; Malik and Godfrey-Faussett 2005; Nicol and Wilkinson 2008; Parwati et al. 2010). One of the most comprehensive articles published recently reviewed 100 papers on the topic, and concluded that even though the phenotype of clinical strains of M. tuberculosis undoubtedly differs significantly in vitro and in animal models of infection, if and how these differences are reflected in clinical settings is less clear (Coscolla and Gagneux 2010). Two years later, this view largely still holds. Nevertheless, recent technological advances, in particular large-scale DNA sequencing and other–omics platforms, are now able to approach the subject at an unprecedented scale, with the potential to generate important novel insights. Given the biological complexities in TB, it is too simplistic to think that variable strain behaviour will be the product of just one or few genomic differences. It is much more likely that the various genomic features of a given strain will have combined effects. Such epistatic interactions will be difficult to detect and link to relevant phenotypes, but systems biology approaches might offer a way forward (Comas and Gagneux 2011).
This chapter summarises some of the recent advances in our understanding of the nature and consequence of strain variation in M. tuberculosis, with a special focus on comparative whole-genome sequencing. I will start by reviewing the current definition of the M. tuberculosis complex (MTBC), and briefly discuss the main genotyping methods for MTBC as well as their limitations. I will then review our current understanding of the global phylogeography of human-associated MTBC based on the available whole-genome data, discuss some of the new forms of genomic variation that have become evident through comparative genome sequencing, and review recent examples where large-scale DNA sequence data was used to predict putative phenotypic effects. Moving from the genome diversity to phenotypic variation, I will review some recent insights into strain-specific transcriptomes and strain-specific differences in innate immune recognition. Finally, I will discuss recent findings on the microevolution of drug-resistant MTBC, and end with a few thoughts on possible future research directions.
2 Genomic Diversity Among Human-Associated MTBC
2.1 The Current Definition of MTBC
MTBC consists of several closely related species and sub-species of acid-fast bacteria also referred to as ‘ecotypes’ (Smith et al. 2005). Even though these ecotypes share identical 16S ribosomal RNA sequences and 99.9 % nucleotide identity at the whole-genome level (with the exception of Mycobacterium canettii and the other so-called “smooth tubercle bacilli” further discussed below), they appear to be adapted to different host species. M. tuberculosis sensu stricto and Mycobacterium africanum are the main agents of TB in humans. M. africanum is limited to West Africa for reasons that are unknown, but causes up to 50 % of human TB in parts of that region (de Jong et al. 2010). In addition to these human pathogens, several animal-associated members of MTBC are found in various domestic and wild animals. These include Mycobacterium bovis, the agent of bovine TB (Garnier et al. 2003), Mycobacterium caprae (sheep and goats) (Niemann et al. 2002), Mycobacterium microti (voles) (Frota et al. 2004), Mycobacterium pinnipedii (seals and see lions) (Cousins et al. 2003), Mycobacterium mungi (mangoose) (Alexander et al. 2010), Mycobacterium orygis (antelope) (van Ingen et al. 2012) and the “dassie bacillus” (rock hyrax) (Mostowy et al. 2004a).
M. canettii and the other smooth TB bacilli (also referred to as “Mycobacterium prototuberculosis”) are regarded by some as human-adapted pathogens (Gutierrez et al. 2005), but several factors point to a likely environmental reservoir for these microbes (Koeck et al. 2010). First, only about 60 isolates have been described in the literature since the original discovery of M. canettii in 1969 (van Soolingen et al. 1997). Second, the majority of these organisms have been isolated from patients in Djibouti (Fabre et al. 2010). Third, no human-to-human transmission has been documented thus far (Koeck et al. 2010). And forth, these bacteria show clear evidence of extensive ongoing horizontal gene transfer (HGT) (Gutierrez et al. 2005), which stands in contrast to the other members of MTBC (the role of HGT and recombination in MTBC is further discussed below).
For the remainder of this chapter, I will focus on M. tuberculosis sensu stricto and M. africanum , which are the most important agents of human TB. Based on the various strain genotyping techniques, M. tuberculosis and M. africanum have been further subdivided into strain lineages and families. For example, the “Beijing” family of strains was originally defined based on characteristic IS6110 RFLP patterns (van Soolingen et al. 1995). This strain family has gained considerable attention because of its association with drug resistance (reviewed in Borrell and Gagneux 2009) and hypervirulence in some experimental models (reviewed in Parwati et al. 2010). Other human-associated MTBC lineages have been studied less, partially because of the lack of a standardised and phylogenetically robust classification system and associated nomenclature (Gagneux and Small 2007). Ultimately, such a system should be based on whole-genome sequencing of a large and representative collection of MTBC clinical strains. However, until genome-sequencing becomes more readily available, the different genotyping methodologies and the corresponding nomenclatures are likely to continue to be used in parallel (for a comparison of the various nomenclatures for human-associated MTBC refer to Coscolla and Gagneux 2010). Let me thus briefly review these different genotyping techniques, and discuss some of their advantages and disadvantages.
2.2 Current Genotyping Methodologies for MTBC
Strain typing in MTBC is usually performed for one of three main purposes: (i) “classical” molecular epidemiology , (ii) phylogenetic and evolutionary studies and (iii) strain classification . However, the existing genotyping techniques are not equally suited for all of these applications. Classical molecular epidemiological studies in TB usually involve measuring ongoing transmission, differentiating between relapse and re-infection, or detecting laboratory cross-contamination (Kato-Maeda et al. 2011). Such studies require highly discriminatory genotyping tools such as IS6110 RFLP analysis (van Embden et al. 1993) or MIRU-VNTR typing (Supply et al. 2006). These techniques rely on mobile and repetitive DNA elements, respectively, which exhibit a rapid rate of change (i.e. a fast “molecular clock”), providing a high discriminatory power for differentiating between patient isolates. However, whilst useful for genotyping closely related strains, these high rates of change lead to convergent evolution and homoplasy (i.e., emergence of identical patterns in phylogenetically unrelated strains), which complicates phylogenetic inference and strain classification (Comas et al. 2009). Spoligotyping is another genotyping technique which has been widely used for both molecular epidemiology and evolutionary studies. It usually shows a lower discriminatory power than IS6110 RFLP or MIRU-VNTR typing, and is therefore often used as a complementary genotyping method in epidemiological studies (Kamerbeek et al. 1997). Spoligotyping is based on the CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) region, also known as the Direct Repeat (DR) region of MTBC. Unique so-called spacer sequences are interspersed between repetitive sequences and are variably present or absent in a given MTBC isolate. Interrogation of 43 of these unique spacer sequences results in a strain-specific fingerprint which is easily digitalised and compared across laboratories. Several large international databases have been compiled containing spoligotypes of thousands of clinical isolates from many countries (Allix-Beguec et al. 2008; Demay et al. 2012). Based on some characteristic spoligopatterns, several MTBC lineages and families have been defined. For example, the Beijing family shows a typical spoligopattern in which the first 34 unique spacers are deleted (Bifani et al. 2002). Unfortunately, spoligotyping patterns, too, are prone to homoplasy as individual spacers can be deleted independently in phylogenetically unrelated strains (Comas et al. 2009). For example, a recent study reported clinical strains exhibiting a “pseudo-Beijing” spoligotype, referring to the fact that these strains showed a spoligotype pattern characteristic of Beijing strains but belonged to a different phylogenetic lineage (Fenner et al. 2011). Moreover, some spoligotyping patterns do not exhibit any of the recognised signatures and can therefore not be classified into any of the known strain families (Flores et al. 2007).
Two additional genotyping approaches have been developed primarily for phylogenetic studies and strain classification in MTBC; one is based on genomic deletions (also referred to as large sequence polymorphisms (LSPs) or regions of difference (RDs)), and the other on single nucleotide polymorphisms (SNPs). Starting about 12 years ago, scientists have used bacterial artificial chromosome libraries and DNA microarrays to interrogate the genome of MTBC strains (Behr et al. 1999; Gordon et al. 1999; Kato-Maeda et al. 2001b; Tsolaki et al. 2004). These studies identified many genomic regions that were absent compared to the H37Rv reference genome, which at the time was the only whole-genome sequence of MTBC available (Cole et al. 1998). Many of these genomic deletions can be used as robust phylogenetic markers for assigning MTBC strain into meaningful groupings. This is because due to the virtual absence of large-scale HGT (further discussed below), once a particular genomic region is lost in MTBC, this region cannot be reacquired, and all progeny of the strain experiencing the loss initially will inherit this deletion. The observation that phylogenetic trees constructed using such genomic deletion markers showed no homoplasy was per se strong support for the already established notion that MTBC exhibits no ongoing HGT and a highly clonal population structure. Several groups used such an approach to revisit the evolutionary history of MTBC and define sets of discrete strain lineages (Brosch et al. 2002; Gagneux et al. 2006b; Hirsh et al. 2004; Mostowy et al. 2002, 2004b; Tsolaki et al. 2005). From a strict phylogenetic point of view; however, these evolutionary scenarios do not represent actual phylogenetic trees but mere cladograms, as their branches do not reflect actual evolutionary distances (i.e., genomic deletions are stochastic events for which no evolutionary models are available). Moreover, phylogenetic inference using genomic deletions is limited by the fact that these markers are usually defined through a one-way comparison with the H37Rv reference genome.
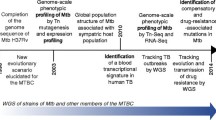
SNP typing is rapidly gaining importance for MTBC genotyping, primarily because of available technologies originally developed for other organisms. It is important to note however that, similarly to genomic deletion analysis, SNP typing does not provide the necessary resolution to be applicable in classical molecular epidemiological studies of TB transmission (Kato-Maeda et al. 2011). Nevertheless, SNP typing is ideal for classifying MTBC strains into lineages as it exhibits negligible levels of homoplasy (Comas et al. 2009). Moreover, various typing methodologies have already been proposed (Dos Vultos et al. 2008; Filliol et al. 2006; Gutacker et al. 2006; Mestre et al. 2011). Importantly, in contrast to de novo SNP discovery through DNA sequencing (Baker et al. 2004; Hershberg et al. 2008), mere SNP typing is of limited use in phylogenetic studies because of the problem known as ‘phylogenetic discovery bias’ (Pearson et al. 2004). This bias refers to the fact that if only a few reference genomes are used to identify the SNPs used for subsequent genotyping, the resulting phylogeny will be biased, because the genetic diversity among strains not included in the initial SNP discovery will not be detected. As a consequence, such strains will automatically fall in intermediate positions on the phylogenetic tree, leading to the problems known as ‘branch collapse’ and ‘linear phylogeny’ (Achtman 2008; Alland et al. 2003; Smith et al. 2009). Recently, two new complementary SNP-typing assays have been developed to screen for the main lineages of MTBC (Stucki et al. 2012). In contrast to most other methods available, the SNPs used in these new assays have been identified by comparing many genomes representative of the known global MTBC diversity (Fig. 1).
2.3 From Genotypes to Whole Genomes
Whole-genome sequencing offers many advantages over the current methods used for genotyping MTBC. The main disadvantage remains the relatively high cost and the bioinformatics capacity required to analyse the data. However, both of these obstacles will likely be surmounted in the near future thanks to further advances in DNA sequencing technologies and the development of more user-friendly analytical tools. Several recent publications support the notion that genome sequencing is already on its way to become the new gold standard for molecular epidemiological studies of MTBC. Specifically, these studies have shown that genome sequencing has a much higher discriminatory power than the standard genotyping tools, and that clinical strains exhibiting identical genotyping profiles can sometimes harbour extensive genetic diversity, with obvious implications for the interpretation of transmission patterns (Casali et al. 2012; Gardy et al. 2011; Niemann et al. 2009). Comparative genome sequencing has also been used to study closely related strains isolated sequentially from a single patient (Comas et al. 2012; Saunders et al. 2011), from chains of ongoing transmission (Sandegren et al. 2011; Schurch et al. 2009, 2010), or from experimentally infected macaques (Ford et al. 2011), in the hope of learning more about the mechanisms driving the micro-evolution of MTBC. Intriguingly, the study in macaques found that the mutation rate of MTBC in latently infected animals was not significantly different from that of bacteria isolated from macaques with active TB. This suggests that dormant and/or persister cells are also prone to mutational damage, and therefore drug resistance could also be acquired during latent infections (Ford et al. 2011).
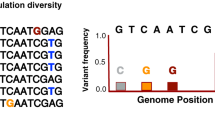
With respect to the global diversity of MTBC, comparative genome sequencing is revealing a degree of heterogeneity which was previously unrecognised. Specifically, because DNA sequence data (as opposed to e.g. genome deletions) can be used to infer genetic distances, this heterogeneity can be measured quantitatively and appreciated in the context of a robust phylogenetic framework. Based on recently published genome sequences (Bentley et al. 2012; Comas et al. 2010), six main phylogenetic lineages can be distinguished among the human-associated MTBC (in addition to all the animal-associated variants not further discussed here; Fig. 1). This includes two lineages traditionally referred to as M. africanum (de Jong et al. 2010). These six lineages have already been described using various genotyping methods and shown to be distributed non-randomly around the world (Fig. 2) (Brudey et al. 2006; Gagneux et al. 2006b). However, as mentioned above, DNA sequence data can be used to compute evolutionary distances and construct robust phylogenetic trees. As shown in Fig. 1, the genetic distance between two human-associated MTBC strains can reach close to 2,000 SNPs, which is equivalent to the evolutionary distance between H37Rv and M. bovis (Garnier et al. 2003). Moreover, about two-thirds of amino acid coding SNPs in MTBC are non-synonymous, and a large proportion of these nSNPs have been predicted to affect gene function (Hershberg et al. 2008). Many more genomes will be necessary to better define the global diversity of MTBC (Fig. 2). In particular, based on the currently available genomes, geographic coverage is limited, and potential phylogenetic substructures within the main lineages are not well understood (Fig. 1). Moreover, we know from genotyping studies that much more MTBC diversity exists that has not yet been characterised by whole-genome sequencing (Demay et al. 2012; Gagneux et al. 2006b).
In addition to genomic deletions and functional SNPs, recent data suggest that gene duplications might also be an important source of genome plasticity in MTBC. In 2000, Brosch et al. reported, two duplicated regions in M. bovis BCG Pasteur (Brosch et al. 2000). Until recently, genomic duplications were considered rare events limited to a few BCG sub-strains (Leung et al. 2008), possibly reflecting the result of long-term in vitro evolution (Brosch et al. 2007). However, two recent studies reported gene duplications in clinical isolates of M. tuberculosis, including one large-scale duplication of more than 500 Kb (Domenech et al. 2010; Weiner et al. 2012). Intriguingly, these duplications occurred multiple times independently and across different MTBC lineages, suggesting that they were positively selected. Moreover, some of these duplications are instable when strains are grown in vitro, arguing against a phenomenon linked to prolonged in vitro growth, but rather in support of in vivo selection and adaptation. Future studies will help define the extent and significance of genomic duplications in MTBC.
2.4 What is the Role of HGT and Recombination in MTBC?
There is strong evidence that past instances of HGT have contributed to the emergence of MTBC as a successful pathogen (Behr and Gagneux 2011). Specifically, genomic comparisons of MTBC and other mycobacteria have identified many genes that the common ancestor of all MTBC appears to have acquired horizontally (Becq et al. 2007; Rosas-Magallanes et al. 2006; Veyrier et al. 2009). Some of these genes have been implicated in the pathogenesis of TB. Similarly, analysis of housekeeping genes in M. canettii and other smooth TB bacilli demonstrated mosaic structures, suggesting ongoing HGT among these rare and extant members of MTBC (Gutierrez et al. 2005). However, whether ongoing (as opposed to past) HGT plays any role in the evolution of MTBC is controversial (except in the case of M. canettii and the other smooth TB bacilli discussed above). For a long time, the common view was that MTBC exhibited a strictly clonal population structure with little evidence for ongoing HGT. With a few exceptions largely considered anecdotal (Hughes et al. 2002; Liu et al. 2006), most of the available evidence supported this view until recently. This included the observation of a rather stable G + C content across most of the MTBC genome (Cole et al. 1998), strong linkage disequilibrium between minisatellite loci (Supply et al. 2003), congruence of phylogenies derived from many different molecular parkers including genomic deletions (Baker et al. 2004; Filliol et al. 2006; Gagneux et al. 2006b; Gagneux and Small 2007; Gutacker et al. 2006; Hirsh et al. 2004; Wirth et al. 2008), and negligible homoplasy in DNA sequence data (Comas et al. 2009; Hershberg et al. 2008). In particular, the fact that genomic deletions in MTBC can be used as robust phylogenetic markers has been demonstrated multiple times (Brosch et al. 2002; Hirsh et al. 2004; Mostowy et al. 2002), and suggests that once a particular genomic region has been lost in MTBC, the corresponding genes cannot be reacquired via HGT. Moreover, all known drug resistance determinants in MTBC represent de novo acquired chromosomal mutations (Sandgren et al. 2009), indicating that HGT plays no significant role in the emergence of drug resistance in this organism.
A recent study has now challenged the dogma of strict clonality in MTBC (Namouchi et al. 2012). The authors used 24 MTBC genomes and performed a series of analyses designed to detect ongoing HGT. Based on their results, the authors concluded that MTBC does in fact show evidence of recombination, mostly involving short DNA fragments of around 50 bp. This observation is striking, given that in other bacteria, recombination usually involves much longer stretches of DNA (Didelot et al. 2010). This short size of recombination tracts might explain why the genomic deletions used to genotype MTBC, which comprise much larger DNA segments, show no evidence of homoplasy linked to HGT or recombination. The authors found that 150 kb (i.e. about 3 %) of the MTBC genomes showed significant recombination tracts. However, the nature of the putative donor organisms could not be determined, except for some instances were the recombining tracks matched regions in the genome of M. canettii. More work is needed, including larger collections of high-quality genomes, to define the role and relevance of HGT and recombination in MTBC, and to identify the sources and mechanisms of acquisition of foreign DNA in this organism.
2.5 Using DNA Sequence Data to Predict the Impact of Genome Diversity
DNA sequences can also be used to analyse the relative strength and direction of selection acting on these sequences. For example, a recent study used whole-genome sequences from 21 clinical strains covering the global diversity of MTBC to study the genetic diversity of 491 experimentally confirmed human T cell epitopes (Comas et al. 2010). In contrast to other pathogens were immune pressure drives increased antigenic diversity (Deitsch et al. 2009), the authors found that in MTBC, T cell epitopes were evolutionarily hyperconserved, showing the lowest dN/dS values (i.e. ratio of non-synonymous to synonymous mutations) in the genome (Fig. 3), with more than 95 % of these epitopes harbouring no amino acid substitution at all. The authors concluded from their findings that the host immune responses directed towards these hyperconserved T cell epitopes might offer a net benefit to the bacteria rather than to the host, by promoting tissue damage, ultimately leading to enhanced transmission of MTBC. This notion is supported by the fact that patient with cavitary disease are particularly likely to generate secondary cases (Rodrigo et al. 1997). Also, in TB patients co-infected with HIV, CD4 T cell counts are inversely correlated with the likelihood of developing cavitations (Kwan and Ernst 2011). The latter suggests that CD4 T cells directly or indirectly contribute to the formation of cavitations, and that TB patients with low CD4 T cell counts might be less likely to transmit MTBC (Brites and Gagneux 2011; Cruciani et al. 2001).
Human T cell epitopes of MTBC are evolutionarily hyperconserved. The ratios of non-synonymous to synonymous nucleotide substitutions are indicated for each gene category (adapted from Comas et al. 2010)
The observation that known human T cell epitopes in MTBC are evolutionarily hyperconserved does of course not exclude the possibility that other, as yet unknown epitopes might be involved in antigenic variation. Indeed, one of the limitations in the study by Comas et al. was that due to technical reasons, the members of the PE/PPE gene family had to be excluded from the analysis (Brennan and Delogu 2002). Some of these genes have previously been shown to be highly diverse (Talarico et al. 2005, 2008), prompting the view they may be implicated in antigenic variation (Banu et al. 2002). A recent study addressed this possibility by analysing the genetic diversity of various PE/PPE genes across a panel of 40 phylogenetically diverse clinical strains (McEvoy et al. 2012). The authors confirmed that PE/PPE genes were more genetically diverse than other genes, but the observed dN/dS values were in average around 1.0, suggesting these genes evolve neutrally. The latter finding argues against the notion that PE/PPE diversity is a result of immune escape. One of the inherent problems in studying PE/PPE genes is that hardly any information exists as to their specific function(s). Future studies will show whether or not the findings by McEvoy et al. can be extrapolated to other PE/PPE genes.
One of the striking findings of comparative DNA sequencing in MTBC has been the observation that in average about two-thirds of coding SNPs are non-synonymous, thus leading to a change in the encoded amino acid (Comas et al. 2010; Fleischmann et al. 2002). Moreover, in silico analyses have shown that 58 % of nonsynonymous SNPs in MTBC are predicted to affect gene function (Hershberg et al. 2008). Recent data from our laboratory using whole-genome sequences have confirmed that even in the deep phylogenetic branches that define the main lineages of MTBC (Fig. 1), close to 50 % of non-synonymous SNPs are predicted to be functional (unpublished data). Taken together, these data highlight the extent of genetic diversity among MTBC strains and lineages, and suggest that this diversity is likely to have phenotypic consequences.
2.6 From MTBC Genotype to Phenotype
Some evidence that genetic diversity in MTBC does indeed translate into phenotypic diversity comes from studies of gene expression. One early study used DNA microarrays to study the transcriptome of 10 clinical strains and found about 500 genes differentially expressed (Gao et al. 2005). More recently, Homolka et al. studied a panel of 17 MTBC strains covering several of the main phylogenetic lineages of MTBC (Fig. 1) using DNA microarrays, and determined their transcriptional profile when grown in vitro and in resting or activated mouse macrophages (Homolka et al. 2010b). The authors detected both strain-specific and lineage-specific gene expression patterns. These patterns were consistent with the phylogenetic position of the corresponding strains, supporting the association between phylogenetic diversity discussed above and phenotypic variation observed experimentally. Moreover, the authors were able to define a core transcriptome that was common to all strains when grown intracellularly. Considering recent technological advances, including the development of RNA sequencing (RNA-seq), future studies will likely uncover additional MTBC diversity at the level of the genome-wide transcriptome. RNA-seq offers many advantages over DNA microarrays, as it allows for detection of unknown transcripts, including novel regulatory RNAs in a strand-specific manner (Sorek and Cossart 2010). A recent RNA-seq analysis of H37Rv grown in vitro revealed many 5’ and 3’ untranslated regions, antisense transcripts and intergenic small RNAs (Arnvig et al. 2011), supporting a role for non-coding RNA in the biology of MTBC (Arnvig and Young 2012).
In addition to differences in gene expression, MTBC phenotypic diversity has also become evident at the level of the host-pathogen interaction, at least in experimental settings. Many studies have reported MTBC strain-specific differences in immunogenicity and virulence using various models of infection; these have been reviewed in detail in (Coscolla and Gagneux 2010). However, only a few studies have been able to link the observed immune- or virulence-phenotypes to particular molecular characteristics of the infecting MTBC strain (Manca et al. 1999; Newton et al. 2006; Reed et al. 2004). In a recent study, Portevin et al. managed to link strain-specific immune-phenotypes with the phylogenetic classification of these strains using a human monocyte-derived macrophage infection model (Portevin et al. 2011). Specifically, the authors infected macrophages from multiple donors with one of 28 strains of MTBC and measured the cytokine profiles at several time points. They found that these strains varied widely in their stimulation of inflammatory cytokines, suggesting that human macrophages perceive and react to these strains very differently (Fig. 4). Despite this wide range of immune responses, the authors detected statistically significant differences between the different MTBC lineages, suggesting that the phylogenetic diversity of MTBC is also reflected at the host-pathogen interface.
IL-6 response in human monocyte-derived macrophages infected with 28 different MTBC strains. Each strain was tested in eight different human donors (adapted from Portevin et al. 2011)
For a long time, one of the main challenges in the field has been linking strain phenotypes defined in the laboratory to relevant clinical phenotypes; so far no consistent picture has emerged (Coscolla and Gagneux 2010). Perhaps one of the most promising avenues has been the study of M. africanum West Africa 2 (i.e. MTBC Lineage 6 in Fig. 1), which causes up to 50 % of TB in some countries of West African (de Jong et al. 2010). Recently, it was shown that Lineage 6 was attenuated in mice compared to H37Rv (Bold et al. 2012). Consistent with these laboratory findings, individuals infected with Lineage 6 strains were less likely to progress towards active TB compared to individuals infected with other strains, even though no difference was observed at the level of transmission (de Jong et al. 2008). Moreover, Lineage 6 has been associated with HIV co-infection in the Gambia, suggesting Lineage 6 might behave as an opportunistic pathogen (de Jong et al. 2005). No such association was observed in Ghana (Meyer et al. 2008), suggesting that even within a particular MTBC lineage, additional phylogeographic variation exists that might be clinically relevant.
3 Micro-Evolution of Drug-Resistant MTBC
A particular aspect of genetic diversity in MTBC that is of obvious clinical relevance relates to drug resistance. MTBC strains resistant to an ever increasing number of antibiotics are threatening global TB control (Gandhi et al. 2010). Over the last years, many of the molecular mechanisms leading to resistance have been elucidated for many of the first- and second-line drugs (Zhang and Yew 2009). Similarly, many of the mutations causing resistance have been identified (Sandgren et al. 2009). What is much less understood is how drug-resistant MTBC strains evolve over the longer run (Borrell and Gagneux 2009). In particular, little is known on how different drug resistance-conferring mutations might interact with each other, with pre-existing strain-specific mutations, or other mutations acquired subsequent to the resistance mutations (Borrell and Gagneux 2011). A particular class of mutations represented by the latter are referred to as compensatory mutations.
3.1 Compensatory Evolution
Acquisition of drug resistance determinants in bacteria is often associated with reduced Darwinian fitness in absence of the drug (Andersson and Levin 1999). However, this fitness cost can be mitigated by subsequent mutations at secondary sites, a phenomenon known as compensatory evolution. Compensation has been described in many bacterial species and antibiotics (reviewed in Andersson and Hughes 2010). In MTBC, a widely cited example has been mutations in the promoter of ahpC leading to over-expression of this gene, believed to compensate for the loss of the katalase-peroxidase (KatG) activity in isoniazid-resistant strains (Sherman et al. 1996). KatG activates isoniazid but also protects the bacterial cell against host-mediated oxidative stress. Over-expression of ahpC, which encodes an alkyl hydroperoxide reductase is thought to partially protect against excess oxidative damage in strains lacking KatG activity. However, the actual role of ahpC promotor mutations in isoniazid resistance remains unclear (Heym et al. 1997). Moreover, molecular epidemiological studies have found these mutations to be rare in clinical settings (Gagneux et al. 2006a), suggesting they play a minor role in the epidemiology of drug-resistant MTBC. Similarly, a compensatory mechanism in the 16S ribosomal RNA of MTBC strains resistant to aminoglycosides was reported (Shcherbakov et al. 2010). However, the corresponding mutation is only rarely observed in clinical settings (Georghiou et al. 2012). By contrast, Comas et al. described a set of novel compensatory mutations in rpoA and rpoC, which encode the alpha- and beta-prime subunits of the RNA polymerase (Comas et al. 2012). These mutations occurred exclusively in rifampicin-resistant strains with resistance-conferring mutations in rpoB, and were associated with an increased competitive fitness in vitro. Moreover, they occurred in up to 30 % of clinical MDR strains, and were significantly over-represented in MDR strains from countries known to suffer from a high burden of MDR-TB (Fig. 5). This suggests that these compensatory mutations contribute to the spread of drug-resistant MTBC strains. Subsequently, the same mutations were reported from XDR strains from Russia (Casali et al. 2012), and in experimentally evolved Salmonella (Brandis et al. 2012). While the mechanism(s) leading to compensation are still unknown, genetic reconstruction of some these rpoA and rpoC mutations in Salmonella showed that they were directly responsible for the improved growth rate of strains carrying rifampicin resistance-conferring mutations in rpoB (Brandis et al. 2012).
Compensatory evolution in rifampicin-resistant MTBC. The proportion of MDR strains carrying compensatory mutations (CMs) in rpoA or rpoC is indicated for strains isolated globally or from countries with a high MDR-TB burden. Light bars indicate mutations very likely to have a compensatory role; black bars indicate all putative compensatory mutations identified (adapted from Comas et al. 2012)
3.2 The Role of the Strain Genetic Background
The interaction between a drug resistance mutation and a compensatory mutation is an example of epistasis (Borrell and Gagneux 2011). Epistatic interaction can generally be defined as a case in which the phenotypic effect of one mutation is modified by the presence or absence of another mutation (Phillips 2008). In addition to epistatic interactions represented by compensatory evolution, interactions between different drug-resistance mutations have also been described. Importantly, work in Escherichia coli and Pseudomonas aeruginosa has shown that the fitness cost associated with a mutation conferring resistance to one drug can be ameliorated by a mutation conferring resistance to a second drug (Trindade et al. 2009; Ward et al. 2009). This phenomenon is referred to as sign epistasis, as the “sign” of the fitness effect associated with the second mutation switches from “negative” to “positive” (Weinreich et al. 2005). Currently, we do not know whether sign epistasis plays any role in the evolution of MDR-TB, but work is needed to explore this disturbing possibility.
There is increasing evidence that the strain genetic background can influence the evolutionary trajectory of drug-resistant MTBC (Borrell and Gagneux 2011). Presumably, this is due to epistatic interactions between pre-existing strain-specific genomic features, and subsequently acquired drug-resistance conferring mutations and compensatory mutations (Mueller et al. 2012). Experimental work in E. coli has shown that such interactions can limit the available mutational pathway(s) towards drug resistance (Toprak et al. 2012). In MTBC, one particular phylogenetic lineage known as “Beijing” has repeatedly been associated with drug resistance in clinical settings (reviewed in Borrell and Gagneux 2009). The underlying basis of this phenomenon remains unknown, but several hypotheses have been proposed (Parwati et al. 2010). For example, based on various non-ysynonymous mutations found in DNA repair genes of Beijing strains, it was hypothesised that these strains might exhibit an intrinsically elevated mutation rate, leading to an enhanced likelihood of acquiring resistance mutations (Dos Vultos et al. 2008). However, fluctuation assays have yielded conflicting results with respect to the in vitro mutation rate of Beijing compared to other MTBC strains (de Steenwinkel et al. 2012; Werngren and Hoffner 2003).
Several studies have reported associations between particular drug resistance-conferring mutations and MTBC lineages, suggesting that the strain genetic background, as defined by MTBC lineage, can influence the mutational pathway to resistance (Gagneux et al. 2006a; Homolka et al. 2010a). For example in isoniazid-resistant MTBC, inhA promoter regions, which are one of the main causes of isoniazid resistance in clinical settings (Zhang and Yew 2009), have been associated with Lineage 1 of MTBC (Fig. 1). The fact that this association was observed in three independent studies supports a biological basis for this phenomenon (Baker et al. 2004; Fenner et al. 2012; Gagneux et al. 2006a). Moreover, Fenner et al. showed recently that putative epistatic interactions between specific isoniazid resistance-conferring mutations and different strain genetic backgrounds can influence the level of resistance to isoniazid in vitro (Fenner et al. 2012).
4 Conclusions
The past few years have witnessed much progress in our understanding and appreciation of strain-to-strain diversity in MTBC. This has partially been a consequence of new opportunities arising from access to novel technologies, including DNA microarrays, and more recently next-generation DNA sequencing (Comas and Gagneux 2009). Fresh insights have also been gained by learning from work in other bacteria. Fifteen years ago, human-associated MTBC was essentially regarded as a “clone”, and strain variation was generally considered irrelevant by the majority of the basic science community (Kato-Maeda et al. 2001a). Today, we know that MTBC harbours more genetic diversity than previously realised, and experimental studies show very clearly that part of this diversity translates into important phenotypic variation (Coscolla and Gagneux 2010). Yet, the role of this diversity for clinical TB remains largely elusive, partially because of the difficulty of linking MTBC strain genotypes to relevant clinical phenotypes. In addition to differences at the DNA level, MTBC strains have been shown to differ at the transcriptome level (Homolka et al. 2010b). Yet, nothing is known with respect to strain variation at the level of regulatory RNAs, which are highly abundant in this organism (Arnvig et al. 2011). Similarly, epigenetics is a rapidly emerging field that has recently been proposed as an avenue to discover host-derived biomarkers for TB (Esterhuyse et al. 2012). However, little work has been done on epigenetics in MTBC, and it is unknown whether strain variability will be reflected at the epigenetic level as well. Much effort today is dedicated to the development of new diagnostics, drugs and vaccines against TB (Young et al. 2008). Based on the existing evidence from other bacterial pathogens, strain genetic diversity in MTBC should be considered during the development of new tools and strategies to better control TB (Gagneux and Small 2007). Increasingly, systems approaches should be used (Comas and Gagneux 2011), not only to determine if and how diversity in MTBC matters for disease control, but to better understand the biology and epidemiology of one of the most important human diseases.
References
Achtman M (2008) Evolution, population structure, and phylogeography of genetically monomorphic bacterial pathogens. Annu Rev Microbiol 62:53–70
Alexander KA, Laver PN, Michel AL, Williams M, van Helden PD, Warren RM, Gey van Pittius NC (2010) Novel Mycobacterium tuberculosis complex pathogen, M. mungi. Emerg Infect Dis 16:1296–1299
Alland D, Whittam TS, Murray MB, Cave MD, Hazbon MH, Dix K, Kokoris M, Duesterhoeft A, Eisen JA, Fraser CM, Fleischmann RD (2003) Modelling bacterial evolution with comparative-genome-based marker systems: application to Mycobacterium tuberculosis evolution and pathogenesis. J Bacteriol 185:3392–3399
Allix-Beguec C, Harmsen D, Weniger T, Supply P, Niemann S (2008) Evaluation and strategy for use of MIRU-VNTRplus, a multifunctional database for online analysis of genotyping data and phylogenetic identification of Mycobacterium tuberculosis complex isolates. J Clin Microbiol 46:2692–2699
Andersson DI, Hughes D (2010) Antibiotic resistance and its cost: is it possible to reverse resistance? Nat Rev Microbiol 8:260–271
Andersson DI, Levin BR (1999) The biological cost of antibiotic resistance. Curr Opin Microbiol 2:489–493
Arnvig KB, Young D (2012) Non-coding RNA and its potential role in Mycobacterium tuberculosis pathogenesis. RNA Biol 9:427–436
Arnvig KB, Comas I, Thomson NR, Houghton J, Boshoff HI, Croucher NJ, Rose G, Perkins TT, Parkhill J, Dougan G, Young DB (2011) Sequence-based analysis uncovers an abundance of non-coding RNA in the total transcriptome of Mycobacterium tuberculosis. PLoS Pathog 7:e1002342
Baker L, Brown T, Maiden MC, Drobniewski F (2004) Silent nucleotide polymorphisms and a phylogeny for Mycobacterium tuberculosis. Emerg Infect Dis 10:1568–1577
Banu S, Honore N, Saint-Joanis B, Philpott D, Prevost MC, Cole ST (2002) Are the PE-PGRS proteins of Mycobacterium tuberculosis variable surface antigens? Mol Microbiol 44:9–19
Becq J, Gutierrez MC, Rosas-Magallanes V, Rauzier J, Gicquel B, Neyrolles O, Deschavanne P (2007) Contribution of horizontally acquired genomic islands to the evolution of the tubercle bacilli. Mol Biol Evol 24:1861–1871
Behr M, Gagneux S (2011) The rise and fall of the Mycobacterium tuberculosis complex. In: Tibayrenc M (ed) Genetics and evolution of infectious diseases. Elsevier, London, pp 651–667
Behr MA, Wilson MA, Gill WP, Salamon H, Schoolnik GK, Rane S, Small PM (1999) Comparative genomics of BCG vaccines by whole-genome DNA microarray. Science 284:1520–1523
Bentley SD, Comas I, Bryant JM, Walker D, Smith NH, Harris SR, Thurston S, Gagneux S, Wood J, Antonio M, Quail MA, Gehre F, Adegbola RA, Parkhill J, de Jong BC (2012) The genome of Mycobacterium africanum West African 2 reveals a lineage-specific locus and genome erosion common to the M. tuberculosis complex. PLoS Negl Trop Dis 6:e1552
Bifani PJ, Mathema B, Kurepina NE, Kreiswirth BN (2002) Global dissemination of the Mycobacterium tuberculosis W-Beijing family strains. Trends Microbiol 10:45–52
Bold TD, Davis DC, Penberthy KK, Cox LM, Ernst JD, de Jong BC (2012) Impaired fitness of Mycobacterium africanum despite secretion of ESAT-6. J Infect Dis 205:984–990
Borrell S, Gagneux S (2009) Infectiousness, reproductive fitness and evolution of drug-resistant Mycobacterium tuberculosis. Int J Tuberc Lung Dis 13:1456–1466
Borrell S, Gagneux S (2011) Strain diversity, epistasis and the evolution of drug resistance in Mycobacterium tuberculosis. Clin Microbiol Infect 17:815–820
Brandis G, Wrande M, Liljas L, Hughes D (2012) Fitness-compensatory mutations in rifampicin-resistant RNA polymerase. Mol Microbiol 85:142–151
Brennan MJ, Delogu G (2002) The PE multigene family: a ‘molecular mantra’ for mycobacteria. Trends Microbiol 10:246–249
Brites D, Gagneux S (2011) Old and new selective pressures on Mycobacterium tuberculosis. Infect Genet Evol 12:678–685
Brosch R, Gordon SV, Buchrieser C, Pym AS, Garnier T, Cole ST (2000) Comparative genomics uncovers large tandem chromosomal duplications in Mycobacterium bovis BCG Pasteur. Yeast 17:111–123
Brosch R, Gordon SV, Marmiesse M, Brodin P, Buchrieser C, Eiglmeier K, Garnier T, Gutierrez C, Hewinson G, Kremer K, Parsons LM, Pym AS, Samper S, van Soolingen D, Cole ST (2002) A new evolutionary scenario for the Mycobacterium tuberculosis complex. Proc Natl Acad Sci USA 99:3684–3689
Brosch R, Gordon SV, Garnier T, Eiglmeier K, Frigui W, Valenti P, Dos Santos S, Duthoy S, Lacroix C, Garcia-Pelayo C, Inwald JK, Golby P, Nunez Garcia J, Hewinson RG, Behr MA, Quail MA, Churcher C, Barrell BG, Parkhill J, Cole ST (2007) Genome plasticity of BCG and impact on vaccine efficacy. Proc Natl Acad Sci USA
Brudey K, Driscoll JR, Rigouts L, Prodinger WM, Gori A, Al-Hajoj SA, Allix C, Aristimuno L, Arora J, Baumanis V, Binder L, Cafrune P, Cataldi A, Cheong S, Diel R, Ellermeier C, Evans JT, Fauville-Dufaux M, Ferdinand S, Garcia de Viedma D, Garzelli C, Gazzola L, Gomes HM, Gutierrez MC, Hawkey PM, van Helden PD, Kadival GV, Kreiswirth BN, Kremer K, Kubin M, Kulkarni SP, Liens B, Lillebaek T, Ly HM, Martin C, Mokrousov I, Narvskaia O, Ngeow YF, Naumann L, Niemann S, Parwati I, Rahim MZ, Rasolofo-Razanamparany V, Rasolonavalona T, Rossetti ML, Rusch-Gerdes S, Sajduda A, Samper S, Shemyakin I, Singh UB, Somoskovi A, Skuce R, van Soolingen D, Streicher EM, Suffys PN, Tortoli E, Tracevska T, Vincent V, Victor TC, Warren R, Yap SF, Zaman K, Portaels F, Rastogi N, Sola C (2006) Mycobacterium tuberculosis complex genetic diversity : mining the fourth international spoligotyping database (SpolDB4) for classification, population genetics and epidemiology. BMC Microbiol 6:23
Casali N, Nikolayevskyy V, Balabanova Y, Ignatyeva O, Kontsevaya I, Harris SR, Bentley SD, Parkhill J, Nejentsev S, Hoffner SE, Horstmann RD, Brown T, Drobniewski F (2012) Microevolution of extensively drug-resistant tuberculosis in Russia. Genome Res 22:735–745
Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE 3rd, Tekaia F, Badcock K, Basham D, Brown D, Chillingworth T, Connor R, Davies R, Devlin K, Feltwell T, Gentles S, Hamlin N, Holroyd S, Hornsby T, Jagels K, Barrell BG et al (1998) Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature 393:537–544
Comas I, Gagneux S (2009) The past and future of tuberculosis research. PLoS Pathog 5:e1000600
Comas I, Gagneux S (2011) A role for systems epidemiology in tuberculosis research. Trends Microbiol 19:492–500
Comas I, Homolka S, Niemann S, Gagneux S (2009) Genotyping of genetically monomorphic bacteria: DNA sequencing in Mycobacterium tuberculosis highlights the limitations of current methodologies. PLoS ONE 4:e7815
Comas I, Chakravartti J, Small PM, Galagan J, Niemann S, Kremer K, Ernst JD, Gagneux S (2010) Human T cell epitopes of Mycobacterium tuberculosis are evolutionarily hyperconserved. Nat Genet 42:498–503
Comas I, Borrell S, Roetzer A, Rose G, Malla B, Kato-Maeda M, Galagan J, Niemann S, Gagneux S (2012) Whole-genome sequencing of rifampicin-resistant Mycobacterium tuberculosis strains identifies compensatory mutations in RNA polymerase genes. Nat Genet 44:106–110
Coscolla M, Gagneux S (2010) Does M. tuberculosis genomic diversity explain disease diversity? Drug Discov Today Dis Mech 7:e43–e59
Cousins DV, Bastida R, Cataldi A, Quse V, Redrobe S, Dow S, Duignan P, Murray A, Dupont C, Ahmed N, Collins DM, Butler WR, Dawson D, Rodriguez D, Loureiro J, Romano MI, Alito A, Zumarraga M, Bernardelli A (2003) Tuberculosis in seals caused by a novel member of the Mycobacterium tuberculosis complex: Mycobacterium pinnipedii sp. nov. Int J Syst Evol Microbiol 53:1305–1314
Cruciani M, Malena M, Bosco O, Gatti G, Serpelloni G (2001) The impact of human immunodeficiency virus type 1 on infectiousness of tuberculosis: a meta-analysis. Clin Infect Dis 33:1922–1930
de Jong BC, Hill PC, Brookes RH, Otu JK, Peterson KL, Small PM, Adegbola RA (2005) Mycobacterium africanum: a new opportunistic pathogen in HIV infection? Aids 19:1714–1715
de Jong BC, Hill PC, Aiken A, Awine T, Antonio M, Adetifa IM, Jackson-Sillah DJ, Fox A, Deriemer K, Gagneux S, Borgdorff MW, McAdam KP, Corrah T, Small PM, Adegbola RA (2008) Progression to active tuberculosis, but not transmission, varies by Mycobacterium tuberculosis lineage in The Gambia. J Infect Dis 198:1037–1043
de Jong BC, Antonio M, Gagneux S (2010) Mycobacterium africanum–review of an important cause of human tuberculosis in West Africa. PLoS Negl Trop Dis 4:e744
de Steenwinkel JE, ten Kate MT, de Knegt GJ, Kremer K, Aarnoutse RE, Boeree MJ, Verbrugh HA, van Soolingen D, Bakker-Woudenberg IA (2012) Drug susceptibility of Mycobacterium tuberculosis Beijing genotype and association with MDR TB. Emerg Infect Dis 18:660–663
Deitsch KW, Lukehart SA, Stringer JR (2009) Common strategies for antigenic variation by bacterial, fungal and protozoan pathogens. Nat Rev Microbiol 7:493–503
Demay C, Liens B, Burguiere T, Hill V, Couvin D, Millet J, Mokrousov I, Sola C, Zozio T, Rastogi N (2012) SITVITWEB—a publicly available international multimarker database for studying Mycobacterium tuberculosis genetic diversity and molecular epidemiology. Infect Genet Evol 12:755–766
Didelot X, Lawson D, Darling A, Falush D (2010) Inference of homologous recombination in bacteria using whole-genome sequences. Genetics 186:1435–1449
Domenech P, Kolly GS, Leon-Solis L, Fallow A, Reed MB (2010) A massive gene duplication event amongst clinical isolates of the Mycobacterium tuberculosis W/Beijing family. J Bacteriol 192:4562–4570
Dos Vultos T, Mestre O, Rauzier J, Golec M, Rastogi N, Rasolofo V, Tonjum T, Sola C, Matic I, Gicquel B (2008) Evolution and diversity of clonal bacteria: the paradigm of Mycobacterium tuberculosis. PLoS ONE 3:e1538
Esterhuyse MM, Linhart HG, Kaufmann SH (2012) Can the battle against tuberculosis gain from epigenetic research? Trends Microbiol 20:220–226
Fabre M, Hauck Y, Soler C, Koeck JL, van Ingen J, van Soolingen D, Vergnaud G, Pourcel C (2010) Molecular characteristics of “Mycobacterium canettii” the smooth Mycobacterium tuberculosis bacilli. Infect Genet Evol 10:1165–1173
Fenner L, Malla B, Ninet B, Dubuis O, Stucki D, Borrell S, Huna T, Bodmer T, Egger M, Gagneux S (2011) “Pseudo-Beijing”: evidence for convergent evolution in the direct repeat region of Mycobacterium tuberculosis. PLoS One 6:e24737
Fenner L, Egger M, Bodmer T, Altpeter E, Zwahlen M, Jaton K, Pfyffer GE, Borrell S, Dubuis O, Bruderer T, Siegrist HH, Furrer H, Calmy A, Fehr J, Stalder JM, Ninet B, Bottger EC, Gagneux S (2012) Effect of mutation and genetic background on drug resistance in Mycobacterium tuberculosis. Antimicrob Agents Chemother 56:3047–3053
Filliol I, Motiwala AS, Cavatore M, Qi W, Hernando Hazbon M, Bobadilla Del Valle M, Fyfe J, Garcia–Garcia L, Rastogi N, Sola C, Zozio T, Guerrero MI, Leon CI, Crabtree J, Angiuoli S, Eisenach KD, Durmaz R, Joloba ML, Rendon A, Sifuentes-Osornio J, Ponce de Leon A, Cave MD, Fleischmann R, Whittam TS, Alland D (2006) Global phylogeny of Mycobacterium tuberculosis based on single nucleotide polymorphism (SNP) analysis: insights into tuberculosis evolution, phylogenetic accuracy of oher DNA fingerprinting systems, and recommendations for a minimal standard SNP set. J Bacteriol 188:759–772
Fleischmann RD, Alland D, Eisen JA, Carpenter L, White O, Peterson J, DeBoy R, Dodson R, Gwinn M, Haft D, Hickey E, Kolonay JF, Nelson WC, Umayam LA, Ermolaeva M, Salzberg SL, Delcher A, Utterback T, Weidman J, Khouri H, Gill J, Mikula A, Bishai W, Jacobs Jr WR, Jr., Venter JC, Fraser CM (2002) Whole-genome comparison of Mycobacterium tuberculosis clinical and laboratory strains. J Bacteriol 184:5479–5490
Flores L, Van T, Narayanan S, Deriemer K, Kato-Maeda M, Gagneux S (2007) Large sequence polymorphisms classify Mycobacterium tuberculosis with ancestral spoligotyping patterns. J Clin Microbiol 45:3393–3395
Ford CB, Lin PL, Chase MR, Shah RR, Iartchouk O, Galagan J, Mohaideen N, Ioerger TR, Sacchettini JC, Lipsitch M, Flynn JL, Fortune SM (2011) Use of whole genome sequencing to estimate the mutation rate of Mycobacterium tuberculosis during latent infection. Nat Genet 43:482–486
Frota CC, Hunt DM, Buxton RS, Rickman L, Hinds J, Kremer K, van Soolingen D, Colston MJ (2004) Genome structure in the vole bacillus, Mycobacterium microti, a member of the Mycobacterium tuberculosis complex with a low virulence for humans. Microbiology 150:1519–1527
Gagneux S, Small PM (2007) Global phylogeography of Mycobacterium tuberculosis and implications for tuberculosis product development. Lancet Infect Dis 7:328–337
Gagneux S, Burgos MV, DeRiemer K, Encisco A, Munoz S, Hopewell PC, Small PM, Pym AS (2006a) Impact of bacterial genetics on the transmission of isoniazid-resistant Mycobacterium tuberculosis. PLoS Pathog 2:e61
Gagneux S, Deriemer K, Van T, Kato-Maeda M, de Jong BC, Narayanan S, Nicol M, Niemann S, Kremer K, Gutierrez MC, Hilty M, Hopewell PC, Small PM (2006b) Variable host-pathogen compatibility in Mycobacterium tuberculosis. Proc Natl Acad Sci USA 103:2869–2873
Gandhi NR, Nunn P, Dheda K, Schaaf HS, Zignol M, van Soolingen D, Jensen P, Bayona J (2010) Multidrug-resistant and extensively drug-resistant tuberculosis: a threat to global control of tuberculosis. Lancet 375:1830–1843
Gao Q, Kripke KE, Saldanha AJ, Yan W, Holmes S, Small PM (2005) Gene expression diversity among Mycobacterium tuberculosis clinical isolates. Microbiology 151:5–14
Gardy JL, Johnston JC, Sui SJ, Cook VJ, Shah L, Brodkin E, Rempel S, Moore R, Zhao Y, Holt R, Varhol R, Birol I, Lem M, Sharma MK, Elwood K, Jones SJ, Brinkman FS, Brunham RC, Tang P (2011) Whole-genome sequencing and social-network analysis of a tuberculosis outbreak. N Engl J Med 364:730–739
Garnier T, Eiglmeier K, Camus JC, Medina N, Mansoor H, Pryor M, Duthoy S, Grondin S, Lacroix C, Monsempe C, Simon S, Harris B, Atkin R, Doggett J, Mayes R, Keating L, Wheeler PR, Parkhill J, Barrell BG, Cole ST, Gordon SV, Hewinson RG (2003) The complete genome sequence of Mycobacterium bovis. Proc Natl Acad Sci USA 100:7877–7882
Georghiou SB, Magana M, Garfein RS, Catanzaro DG, Catanzaro A, Rodwell TC (2012) Evaluation of genetic mutations associated with Mycobacterium tuberculosis resistance to amikacin, kanamycin and capreomycin: a systematic review. PLoS One 7:e33275
Gordon SV, Brosch R, Billault A, Garnier T, Eiglmeier K, Cole ST (1999) Identification of variable regions in the genomes of tubercle bacilli using bacterial artificial chromosome arrays. Mol Microbiol 32:643–655
Gutacker MM, Mathema B, Soini H, Shashkina E, Kreiswirth BN, Graviss EA, Musser JM (2006) Single-nucleotide polymorphism-based population genetic analysis of Mycobacterium tuberculosis strains from 4 geographic sites. J Infect Dis 193:121–128
Gutierrez C, Brisse S, Brosch R, Fabre M, Omais B, Marmiesse M, Supply P, Vincent V (2005) Ancient origin and gene mosaicism of the progenitor of Mycobacterium tuberculosis. PLoS Pathogens 1:1–7
Hershberg R, Lipatov M, Small PM, Sheffer H, Niemann S, Homolka S, Roach JC, Kremer K, Petrov DA, Feldman MW, Gagneux S (2008) High functional diversity in Mycobacterium tuberculosis driven by genetic drift and human demography. PLoS Biol 6:e311
Heym B, Stavropoulos E, Honore N, Domenech P, Saint-Joanis B, Wilson TM, Collins DM, Colston MJ, Cole ST (1997) Effects of overexpression of the alkyl hydroperoxide reductase AhpC on the virulence and isoniazid resistance of Mycobacterium tuberculosis. Infect Immun 65:1395–1401
Hirsh AE, Tsolaki AG, DeRiemer K, Feldman MW, Small PM (2004) Stable association between strains of Mycobacterium tuberculosis and their human host populations. Proc Natl Acad Sci USA 101:4871–4876
Homolka S, Meyer CG, Hillemann D, Owusu-Dabo E, Adjei O, Horstmann RD, Browne EN, Chinbuah A, Osei I, Gyapong J, Kubica T, Ruesch-Gerdes S, Niemann S (2010a) Unequal distribution of resistance-conferring mutations among Mycobacterium tuberculosis and Mycobacterium africanum strains from Ghana. Int J Med Microbiol 300:489–495
Homolka S, Niemann S, Russell DG, Rohde KH (2010b) Functional genetic diversity among Mycobacterium tuberculosis complex clinical isolates: delineation of conserved core and lineage-specific transcriptomes during intracellular survival. PLoS Pathog 6:e1000988
Hughes AL, Friedman R, Murray M (2002) Genomewide pattern of synonymous nucleotide substitution in two complete genomes of Mycobacterium tuberculosis. Emerg Infect Dis 8:1342–1346
Kamerbeek J, Schouls L, Kolk A, van Agterveld M, van Soolingen D, Kuijper S, Bunschoten A, Molhuizen H, Shaw R, Goyal M, van Embden J (1997) Simultaneous detection and strain differentiation of Mycobacterium tuberculosis for diagnosis and epidemiology. J Clin Microbiol 35:907–914
Kato-Maeda M, Bifani PJ, Kreiswirth BN, Small PM (2001a) The nature and consequence of genetic variability within Mycobacterium tuberculosis. J Clin Invest 107:533–537
Kato-Maeda M, Rhee JT, Gingeras TR, Salamon H, Drenkow J, Smittipat N, Small PM (2001b) Comparing genomes within the species Mycobacterium tuberculosis. Genome Res 11:547–554
Kato-Maeda M, Metcalfe JZ, Flores L (2011) Genotyping of Mycobacterium tuberculosis: application in epidemiologic studies. Future Microbiol 6:203–216
Koeck JL, Fabre M, Simon F, Daffe M, Garnotel E, Matan AB, Gerome P, Bernatas JJ, Buisson Y, Pourcel C (2010) Clinical characteristics of the smooth tubercle bacilli “Mycobacterium canettii” infection suggest the existence of an environmental reservoir. Clin Microbiol Infect 17:1013–1019
Kwan CK, Ernst JD (2011) HIV and tuberculosis: a deadly human syndemic. Clin Microbiol Rev 24:351–376
Leung AS, Tran V, Wu Z, Yu X, Alexander DC, Gao GF, Zhu B, Liu J (2008) Novel genome polymorphisms in BCG vaccine strains and impact on efficacy. BMC Genomics 9:413
Liu X, Gutacker MM, Musser JM, Fu YX (2006) Evidence for recombination in Mycobacterium tuberculosis. J Bacteriol 188:8169–8177
Malik AN, Godfrey-Faussett P (2005) Effects of genetic variability of Mycobacterium tuberculosis strains on the presentation of disease. Lancet Infect Dis 5:174–183
Manca C, Tsenova L, Barry CE 3rd, Bergtold A, Freeman S, Haslett PA, Musser JM, Freedman VH, Kaplan G (1999) Mycobacterium tuberculosis CDC1551 induces a more vigorous host response in vivo and in vitro, but is not more virulent than other clinical isolates. J Immunol 162:6740–6746
McEvoy C, Cloete R, Muller B, Schurch A, van Helden P, Gagneux S, Warren R, Van Pittius NC (2012) Comparative analysis of Mycobacterium tuberculosis PE and PPE genes reveals high sequence variation and an apparent absence of selective constraints. PLoS ONE 7:e30593
Mestre O, Luo T, Dos Vultos T, Kremer K, Murray A, Namouchi A, Jackson C, Rauzier J, Bifani P, Warren R, Rasolofo V, Mei J, Gao Q, Gicquel B (2011) Phylogeny of Mycobacterium tuberculosis Beijing strains constructed from polymorphisms in genes involved in DNA replication, recombination and repair. PLoS One 6:e16020
Meyer CG, Scarisbrick G, Niemann S, Browne EN, Chinbuah MA, Gyapong J, Osei I, Owusu-Dabo E, Kubica T, Rusch-Gerdes S, Thye T, Horstmann RD (2008) Pulmonary tuberculosis: virulence of Mycobacterium africanum and relevance in HIV co-infection. Tuberculosis (Edinb) 88:482–489
Mitchison DA, Wallace JG, Bhatia AL, Selkon JB, Subbaiah TV, Lancaster MC (1960) A comparison of the virulence in guinea-pigs of South Indian and British tubercle bacilli. Tubercle 41:1–22
Mostowy S, Cousins D, Brinkman J, Aranaz A, Behr MA (2002) Genomic deletions suggest a phylogeny for the Mycobacterium tuberculosis complex. J Infect Dis 186:74–80
Mostowy S, Cousins D, Behr MA (2004a) Genomic interrogation of the dassie bacillus reveals it as a unique RD1 mutant within the Mycobacterium tuberculosis complex. J Bacteriol 186:104–109
Mostowy S, Onipede A, Gagneux S, Niemann S, Kremer K, Desmond EP, Kato-Maeda M, Behr M (2004b) Genomic analysis distinguishes Mycobacterium africanum. J Clin Microbiol 42:3594–3599
Mueller B, Borrell S, Rose G, Gagneux S (2012) The heterogeneous evolution of multidrug-resistant Mycobacterium tuberculosis. Trends Genet (in press)
Musser JM, Amin A, Ramaswamy S (2000) Negligible genetic diversity of Mycobacterium tuberculosis host immune system protein targets: evidence of limited selective pressure. Genetics 155:7–16
Namouchi A, Didelot X, Schock U, Gicquel B, Rocha EP (2012) After the bottleneck: genome-wide diversification of the Mycobacterium tuberculosis complex by mutation, recombination, and natural selection. Genome Res 22:721–734
Newton SM, Smith RJ, Wilkinson KA, Nicol MP, Garton NJ, Staples KJ, Stewart GR, Wain JR, Martineau AR, Fandrich S, Smallie T, Foxwell B, Al-Obaidi A, Shafi J, Rajakumar K, Kampmann B, Andrew PW, Ziegler-Heitbrock L, Barer MR, Wilkinson RJ (2006) A deletion defining a common Asian lineage of Mycobacterium tuberculosis associates with immune subversion. Proc Natl Acad Sci USA 103:15594–15598
Nicol MP, Wilkinson RJ (2008) The clinical consequences of strain diversity in Mycobacterium tuberculosis. Trans R Soc Trop Med Hyg 102:955–965
Niemann S, Richter E, Rusch-Gerdes S (2002) Biochemical and genetic evidence for the transfer of Mycobacterium tuberculosis subsp. Caprae Aranaz et al. 1999 to the species Mycobacterium bovis Karlson and Lessel 1970 (approved lists 1980) as Mycobacterium bovis subsp. caprae comb. nov. Int J Syst Evol Microbiol 52:433–436
Niemann S, Koser CU, Gagneux S, Plinke C, Homolka S, Bignell H, Carter RJ, Cheetham RK, Cox A, Gormley NA, Kokko-Gonzales P, Murray LJ, Rigatti R, Smith VP, Arends FP, Cox HS, Smith G, Archer JA (2009) Genomic diversity among drug sensitive and multidrug resistant isolates of Mycobacterium tuberculosis with identical DNA fingerprints. PLoS ONE 4:e7407
Parwati I, van Crevel R, van Soolingen D (2010) Possible underlying mechanisms for successful emergence of the Mycobacterium tuberculosis Beijing genotype strains. Lancet Infect Dis 10:103–111
Pearson T, Busch JD, Ravel J, Read TD, Rhoton SD, U’Ren JM, Simonson TS, Kachur SM, Leadem RR, Cardon ML, Van Ert MN, Huynh LY, Fraser CM, Keim P (2004) Phylogenetic discovery bias in Bacillus anthracis using single-nucleotide polymorphisms from whole-genome sequencing. Proc Natl Acad Sci USA 101:13536–13541
Phillips PC (2008) Epistasis–the essential role of gene interactions in the structure and evolution of genetic systems. Nat Rev Genet 9:855–867
Portevin D, Gagneux S, Comas I, Young D (2011) Human macrophage responses to clinical isolates from the Mycobacterium tuberculosis complex discriminate between ancient and modern lineages. PLoS Pathog 7:e1001307
Rajakumar K, Shafi J, Smith RJ, Stabler RA, Andrew PW, Modha D, Bryant G, Monk P, Hinds J, Butcher PD, Barer MR (2004) Use of genome level-informed PCR as a new investigational approach for analysis of outbreak-associated Mycobacterium tuberculosis isolates. J Clin Microbiol 42:1890–1896
Reed MB, Domenech P, Manca C, Su H, Barczak AK, Kreiswirth BN, Kaplan G, Barry CE 3rd (2004) A glycolipid of hypervirulent tuberculosis strains that inhibits the innate immune response. Nature 431:84–87
Rodrigo T, Cayla JA, Garcia de Olalla P, Galdos-Tanguis H, Jansa JM, Miranda P, Brugal T (1997) Characteristics of tuberculosis patients who generate secondary cases. Int J Tuberc Lung Dis 1:352–357
Rosas-Magallanes V, Deschavanne P, Quintana-Murci L, Brosch R, Gicquel B, Neyrolles O (2006) Horizontal transfer of a virulence operon to the ancestor of Mycobacterium tuberculosis. Mol Biol Evol 23:1129–1135
Sandegren L, Groenheit R, Koivula T, Ghebremichael S, Advani A, Castro E, Pennhag A, Hoffner S, Mazurek J, Pawlowski A, Kan B, Bruchfeld J, Melefors O, Kallenius G (2011) Genomic Stability over 9 Years of an isoniazid resistant Mycobacterium tuberculosis outbreak strain in Sweden. PLoS One 6:e16647
Sandgren A, Strong M, Muthukrishnan P, Weiner BK, Church GM, Murray MB (2009) Tuberculosis drug resistance mutation database. PLoS Med 6:e2
Saunders NJ, Trivedi UH, Thomson ML, Doig C, Laurenson IF, Blaxter ML (2011) Deep resequencing of serial sputum isolates of Mycobacterium tuberculosis during therapeutic failure due to poor compliance reveals stepwise mutation of key resistance genes on an otherwise stable genetic background. J Infect 62:212–217
Schurch AC, Kremer K, Kiers A, Daviena O, Boeree MJ, Siezen R, Smith NH, van Soolingen D (2009) The tempo and mode of molecular evolution of Mycobacterium tuberculosis at patient-to-patient scale. Infect Genet Evol 10:108–114
Schurch AC, Kremer K, Daviena O, Kiers A, Boeree MJ, Siezen RJ, van Soolingen D (2010) High resolution typing by integration of genome sequencing data in a large tuberculosis cluster. J Clin Microbiol 48:3403–3406
Shcherbakov D, Akbergenov R, Matt T, Sander P, Andersson DI, Bottger EC (2010) Directed mutagenesis of Mycobacterium smegmatis 16S rRNA to reconstruct the in vivo evolution of aminoglycoside resistance in Mycobacterium tuberculosis. Mol Microbiol 77:830–840
Sherman DR, Mdluli K, Hickey MJ, Arain TM, Morris SL, Barry CE 3rd, Stover CK (1996) Compensatory ahpC gene expression in isoniazid-resistant Mycobacterium tuberculosis. Science 272:1641–1643
Smith NH, Kremer K, Inwald J, Dale J, Driscoll JR, Gordon SV, van Soolingen D, Glyn Hewinson R, Maynard Smith J (2005) Ecotypes of the Mycobacterium tuberculosis complex. J Theor Biol 239:220–225
Smith NH, Hewinson RG, Kremer K, Brosch R, Gordon SV (2009) Myths and misconceptions: the origin and evolution of Mycobacterium tuberculosis. Nat Rev Microbiol 7:537–544
Sorek R, Cossart P (2010) Prokaryotic transcriptomics: a new view on regulation, physiology and pathogenicity. Nat Rev Genet 11:9–16
Sreevatsan S, Pan X, Stockbauer KE, Connell ND, Kreiswirth BN, Whittam TS, Musser JM (1997) Restricted structural gene polymorphism in the Mycobacterium tuberculosis complex indicates evolutionarily recent global dissemination. Proc Natl Acad Sci USA 94:9869–9874
Stucki D, Malla B, Hostettler S, Huna T, Feldmann J, Yeboah-Manu D, Borrell S, Fenner L, Comas I, Coscolla M, Gagneux S (2012) Two new rapid SNP-typing methods for classifying Mycobacterium tuberculosis complex into the main phylogenetic lineages. PLoS ONE 7:e41253
Supply P, Warren RM, Banuls AL, Lesjean S, Van Der Spuy GD, Lewis LA, Tibayrenc M, Van Helden PD, Locht C (2003) Linkage disequilibrium between minisatellite loci supports clonal evolution of Mycobacterium tuberculosis in a high tuberculosis incidence area. Mol Microbiol 47:529–538
Supply P, Allix C, Lesjean S, Cardoso-Oelemann M, Rusch-Gerdes S, Willery E, Savine E, de Haas P, van Deutekom H, Roring S, Bifani P, Kurepina N, Kreiswirth B, Sola C, Rastogi N, Vatin V, Gutierrez MC, Fauville M, Niemann S, Skuce R, Kremer K, Locht C, van Soolingen D (2006) Proposal for standardization of optimized Mycobacterial Interspersed Repetitive Unit-Variable Number Tandem Repeat typing of Mycobacterium tuberculosis. J Clin Microbiol 44:4498–4510
Talarico S, Cave MD, Marrs CF, Foxman B, Zhang L, Yang Z (2005) Variation of the Mycobacterium tuberculosis PE_PGRS 33 gene among clinical isolates. J Clin Microbiol 43:4954–4960
Talarico S, Zhang L, Marrs CF, Foxman B, Cave MD, Brennan MJ, Yang Z (2008) Mycobacterium tuberculosis PE_PGRS16 and PE_PGRS26 genetic polymorphism among clinical isolates. Tuberculosis (Edinb) 88:283–294
Toprak E, Veres A, Michel JB, Chait R, Hartl DL, Kishony R (2012) Evolutionary paths to antibiotic resistance under dynamically sustained drug selection. Nat Genet 44:101–105
Trindade S, Sousa A, Xavier KB, Dionisio F, Ferreira MG, Gordo I (2009) Positive epistasis drives the acquisition of multidrug resistance. PLoS Genet 5:e1000578
Tsolaki AG, Hirsh AE, DeRiemer K, Enciso JA, Wong MZ, Hannan M, Goguet de la Salmoniere YO, Aman K, Kato-Maeda M, Small PM (2004) Functional and evolutionary genomics of Mycobacterium tuberculosis: insights from genomic deletions in 100 strains. Proc Natl Acad Sci USA 101:4865–4870
Tsolaki AG, Gagneux S, Pym AS, Goguet de la Salmoniere YO, Kreiswirth BN, Van Soolingen D, Small PM (2005) Genomic deletions classify the Beijing/W strains as a distinct genetic lineage of Mycobacterium tuberculosis. J Clin Microbiol 43:3185–3191
Valway SE, Sanchez MP, Shinnick TF, Orme I, Agerton T, Hoy D, Jones JS, Westmoreland H, Onorato IM (1998) An outbreak involving extensive transmission of a virulent strain of Mycobacterium tuberculosis. N Engl J Med 338:633–639
van Embden JD, Cave MD, Crawford JT, Dale JW, Eisenach KD, Gicquel B, Hermans P, Martin C, McAdam R, Shinnick TM et al (1993) Strain identification of Mycobacterium tuberculosis by DNA fingerprinting: recommendations for a standardized methodology. J Clin Microbiol 31:406–409
van Ingen J, Rahim Z, Mulder A, Boeree MJ, Simeone R, Brosch R, van Soolingen D (2012) Characterization of Mycobacterium orygis as M. tuberculosis complex subspecies. Emerg Infect Dis 18:653–655
van Soolingen D, Qian L, de Haas PE, Douglas JT, Traore H, Portaels F, Qing HZ, Enkhsaikan D, Nymadawa P, van Embden JD (1995) Predominance of a single genotype of Mycobacterium tuberculosis in countries of east Asia. J Clin Microbiol 33:3234–3238
van Soolingen D, Hoogenboezem T, de Haas PE, Hermans PW, Koedam MA, Teppema KS, Brennan PJ, Besra GS, Portaels F, Top J, Schouls LM, van Embden JD (1997) A novel pathogenic taxon of the Mycobacterium tuberculosis complex, Canetti: characterization of an exceptional isolate from Africa. Int J Syst Bacteriol 47:1236–1245
Veyrier F, Pletzer D, Turenne C, Behr MA (2009) Phylogenetic detection of horizontal gene transfer during the step-wise genesis of Mycobacterium tuberculosis. BMC Evol Biol 9:196
Ward H, Perron GG, Maclean RC (2009) The cost of multiple drug resistance in Pseudomonas aeruginosa. J Evol Biol 22:997–1003
Weiner B, Gomez J, Victor TC, Warren RM, Sloutsky A, Plikaytis BB, Posey JE, van Helden PD, Gey van Pittius NC, Koehrsen M, Sisk P, Stolte C, White J, Gagneux S, Birren B, Hung D, Murray M, Galagan J (2012) Independent large scale duplications in multiple M. tuberculosis lineages overlapping the same genomic region. PLoS One 7:e26038
Weinreich DM, Watson RA, Chao L (2005) Perspective: sign epistasis and genetic constraint on evolutionary trajectories. Evolution 59:1165–1174
Werngren J, Hoffner SE (2003) Drug-susceptible Mycobacterium tuberculosis Beijing genotype does not develop mutation-conferred resistance to rifampin at an elevated rate. J Clin Microbiol 41:1520–1524
Wirth T, Hildebrand F, Allix-Beguec C, Wolbeling F, Kubica T, Kremer K, van Soolingen D, Rusch-Gerdes S, Locht C, Brisse S, Meyer A, Supply P, Niemann S (2008) Origin, spread and demography of the Mycobacterium tuberculosis complex. PLoS Pathog 4:e1000160
Young DB, Perkins MD, Duncan K, Barry CE (2008) Confronting the scientific obstacles to global control of tuberculosis. J Clin Invest 118:1255–1265
Zhang Y, Yew WW (2009) Mechanisms of drug resistance in Mycobacterium tuberculosis. Int J Tuberc Lung Dis 13:1320–1330
Zhang M, Gong J, Yang Z, Samten B, Cave MD, Barnes PF (1999) Enhanced capacity of a widespread strain of Mycobacterium tuberculosis to grow in human macrophages. J Infect Dis 179:1213–1217
Acknowledgments
I thank Douglas Young, Iñaki Comas and all the members of my group for the stimulating discussions and comments on the manuscript. Work in my laboratory is supported by the Swiss National Science Foundation (grant number PP0033-119205), the National Institutes of Health (AI090928 and HHSN266200700022C) the Leverhulme-Royal Society Africa Award (AA080019) and the European Research Council (309540).
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Gagneux, S. (2013). Genetic Diversity in Mycobacterium tuberculosis . In: Pieters, J., McKinney, J. (eds) Pathogenesis of Mycobacterium tuberculosis and its Interaction with the Host Organism. Current Topics in Microbiology and Immunology, vol 374. Springer, Berlin, Heidelberg. https://doi.org/10.1007/82_2013_329
Download citation
DOI: https://doi.org/10.1007/82_2013_329
Published:
Publisher Name: Springer, Berlin, Heidelberg
Print ISBN: 978-3-642-40231-9
Online ISBN: 978-3-642-40232-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)