Abstract
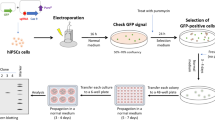
The ability to reprogram somatic cells into induced pluripotent stem cells (iPSCs) was developed in 2006 and represented a major breakthrough in stem cell research. A more recent milestone in biomedical research was reached in 2013 when the CRISPR/Cas9 system was used to edit the genome of mammalian cells. The coupling of both human (h)iPSCs and CRISPR/Cas9 technology offers great promise for cell therapy and regenerative medicine. However, several limitations including time and labor consumption, efficiency and efficacy of the system, and the potential off-targets effects induced by the Cas9 nuclease still need to be addressed. Here, we describe a detailed method for easily engineering genetic changes in hiPSCs, using a nucleofection-mediated protocol to deliver the CRISPR/Cas9 components into the cells, and discuss key points to be considered when designing your experiment. The clonal, genome-edited hiPSC line generated via our method can be directly used for downstream applications.
Access this chapter
Tax calculation will be finalised at checkout
Purchases are for personal use only
Similar content being viewed by others
References
Greenwald DL, Cashman SM, Kumar-Singh R (2010) Engineered zinc finger nuclease-mediated homologous recombination of the human rhodopsin gene. Investig Ophthalmol Vis Sci 51:6374–6380
Overlack N, Goldmann T, Wolfrum U, Nagel-Wolfrum K (2012) Gene repair of an usher syndrome causing mutation by zinc-finger nuclease mediated homologous recombination. Investig Ophthalmol Vis Sci 53:4140–4146
Low BE, Krebs MP, Joung JK, Tsai SQ, Nishina PM, Wiles MV (2014) Correction of the Crb1rd8 allele and retinal phenotype in C57BL/6N mice via TALEN-mediated homology-directed repair. Investig Ophthalmol Vis Sci 55:387–395
Miller JB, Zhang S, Kos P, Xiong H, Zhou K, Perelman SS, Zhu H, Siegwart DJ (2017) Non-viral CRISPR/Cas gene editing in vitro and in vivo enabled by synthetic nanoparticle co-delivery of Cas9 mRNA and sgRNA. Angew Chem Int Ed Engl 56:1059–1063
Hockemeyer D, Wang H, Kiani S, Lai CS, Gao Q, John P, Cost GJ, Zhang L, Santiago Y, Miller JC, Cherone JM, Meng X, Hinkley SJ, Rebar EJ, Gregory D, Urnov FD, Jaenisch R (2012) Genetic engineering of human ES and iPS cells using TALE nucleases. Nat Biotechnol 29:731–734
Sanjurjo-Soriano C, Erkilic N, Baux D, Mamaeva D, Hamel CP, Meunier I, Roux AF, Kalatzis V (2020) Genome editing in patient iPSCs corrects the most prevalent USH2A mutations and reveals intriguing mutant mRNA expression profiles. Mol Ther 17:156–173
Jinek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E (2012) A Programmable Dual-RNA–Guided DNA Endonuclease in Adaptive Bacterial Immunity. Science 337:816–821
Sanjurjo-Soriano C, Kalatzis V (2018) Guiding lights in genome editing for inherited retinal disorders: implications for gene and Cell therapy. Neural Plast 8:5056279
Strouse B, Bialk P, Niamat RA, Rivera-torres N, Kmiec EB (2014) Combinatorial gene editing in mammalian cells using ssODNs and TALENs. Sci Rep 4:3791
Yang L, Guell M, Byrne S, Yang JL, De Los Angeles A, Mali P, Aach J, Kim-Kiselak C, Briggs AW, Rios X, Huang PY, Daley G, Church G (2013) Optimization of scarless human stem cell genome editing. Nucleic Acids Res 41:9049–9061
Paquet D, Kwart D, Chen A, Sproul A, Jacob S, Teo S, Olsen KM, Gregg A, Noggle S, Tessier-Lavigne M (2016) Efficient introduction of specific homozygous and heterozygous mutations using CRISPR/Cas9. Nature 533:125–129
Ding Q, Regan SN, Xia Y, Oostrom LA, Cowan CA, Musunuru K (2013) Enhanced efficiency of human pluripotent stem cell genome editing through replacing TALENs with CRISPRs. Cell Stem Cell 12:393–394
Takahashi K, Yamanaka S (2006) Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell 126:663–676
Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S (2007) Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell 131:861–872
Guenther MG, Frampton GM, Soldner F, Hockemeyer D, Mitalipova M, Jaenisch R, Young RA (2010) Chromatin structure and gene expression programs of human embryonic and induced pluripotent stem cells. Cell Stem Cell 7:249–257
Zhao MT, Chen H, Liu Q, Shao NY, Sayed N, Wo HT, Zhang JZ, Ong SG, Liu C, Kim Y, Yang H, Chour T, Ma H, Gutierrez NM, Karakikes I, Mitalipov S, Snyder MP, Wu JC (2017) Molecular and functional resemblance of differentiated cells derived from isogenic human iPSCs and SCNT-derived ESCs. Proc Natl Acad Sci U S A 114:E11111–E11120
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK (2014) Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol 32:279–284
Concordet JP, Haeussler M (2018) CRISPOR: intuitive guide selection for CRISPR/Cas9 genome editing experiments and screens. Nucleic Acids Res 46:W242–W245
Montague TG, Cruz JM, Gagnon JA, Church GM, Valen E (2014) CHOPCHOP: a CRISPR/Cas9 and TALEN web tool for genome editing. Nucleic Acids Res 42:401–407
Ran FA, Hsu PD, Wright J, Agarwala V, Scott DA, Zhang F (2013) Genome engineering using the CRISPR-Cas9 system. Nat Protoc 8:2281–2308
Bai Q, Ramirez J-M, Becker F, Pantesco V, Lavabre-Bertrand T, Hovatta O, Lemaître J-M, Pellestor F, De Vos J (2015) Temporal analysis of genome alterations induced by single-cell passaging in human embryonic stem cells. Stem Cells Dev 24:653–662
Kyriakides O, Halliwell JA, Andrews PW (2018) Acquired genetic and epigenetic variation in human pluripotent stem cells. Adv Biochem Eng Biotechnol 163:187–206
Richardson CD, Ray GJ, DeWitt MA, Curie GL, Corn JE (2016) Enhancing homology-directed genome editing by catalytically active and inactive CRISPR-Cas9 using asymmetric donor DNA. Nat Biotechnol 34:339–344
Kato-inui T, Takahashi G, Hsu S, Miyaoka Y (2018) Clustered regularly interspaced short palindromic repeats (CRISPR) /CRISPR-associated protein 9 with improved proof-reading enhances homology-directed repair. Nucleic Acids Res 46:4677–4688
Kulcsár PI, Tálas A, Huszár K, Ligeti Z, Tóth E, Weinhardt N, Fodor E, Welker E (2017) Crossing enhanced and high fidelity SpCas9 nucleases to optimize specificity and cleavage. Genome Biol 18:190
Yamamoto Y, Makiyama T, Harita T, Sasaki K, Wuriyanghai Y, Hayano M, Nishiuchi S, Kohjitani H, Hirose S, Chen J, Yokoi F, Ishikawa T, Ohno S, Chonabayashi K, Motomura H, Yoshida Y, Horie M, Makita N, Kimura T (2017) Allele-specific ablation rescues electrophysiological abnormalities in a human iPS cell model of long-QT syndrome with a CALM2 mutation. Hum Mol Genet 26:1670–1677
Acknowledgments
This work was supported by the associations Aviesan-Unadev, France Choroideremia, and Vaincre Usher 2. We thank the MRI imaging facility for FACS analysis.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2021 Springer Science+Business Media, LLC
About this protocol
Cite this protocol
Sanjurjo-Soriano, C., Erkilic, N., Mamaeva, D., Kalatzis, V. (2021). CRISPR/Cas9-Mediated Genome Editing to Generate Clonal iPSC Lines. In: Nagy, A., Turksen, K. (eds) Induced Pluripotent Stem (iPS) Cells. Methods in Molecular Biology, vol 2454. Humana, New York, NY. https://doi.org/10.1007/7651_2021_362
Download citation
DOI: https://doi.org/10.1007/7651_2021_362
Published:
Publisher Name: Humana, New York, NY
Print ISBN: 978-1-0716-2118-9
Online ISBN: 978-1-0716-2119-6
eBook Packages: Springer Protocols