Abstract
Background
Methicillin-resistant Staphylococcus aureus is a global public health challenge and there is a continuous increase in community-acquired infections among people in different geographical location. We sought the distribution and antibiotics pattern of community-acquired methicillin-resistant Staphylococcus isolates among apparently healthy residents of Ibadan, Southwestern Nigeria.
Methods
Seven hundred (700) healthy volunteers residing in Ibadan metropolis, Nigeria, were enrolled in this study. Isolates from the nasal swabs were aseptically collected and characterized using standard and established microbiological methods, which included growth and fermentation on mannitol salt agar, colonial morphology, Gram-staining reaction, Microbact™ 12S identification kit and confirmed with 16SrRNA. After identification of the isolates, antimicrobial susceptibility test was performed on Mueller-Hinton agar by modified Kirby-Bauer disc diffusion method and the presence of mecA and nuc genes were detected via polymerase chain reaction assay.
Results
Prevalence of Staphylococcus aureus nasal carriage and Methicillin-resistant Staphylococcus in this study was 31.9% and 9.43% respectively. The residents of Ibadan North local government area (Fisher’s Exact = 1.8962, P = .028) and Egbeda local government area (Fisher’s Exact = 2.7222, P = .006) are likely to carry Methicillin-resistant Staphylococcus than any other local government area in Ibadan, Nigeria. The antimicrobial resistance patterns of the isolates revealed high resistance to Oxacillin (96.9%). Most of the isolates were sensitive to vancomycin (92.4%). Polymerase chain reaction analysis showed that mecA gene was present in all 66 (100%) Methicillin-resistant Staphylococcus aureus isolates. Male-gender (ϰ2 = 8.849, P = .003), Adults; 40–50 years old (ϰ2 = 9.842, P = .002), low educational background (ϰ2 = 36.817, P ˂ .001), recent hospital visitation (ϰ2 = 8.693, P = .003) are some of the factors that are observed in this study to be associated with Methicillin-resistant Staphylococcus infection.
Conclusion
Our findings revealed the relatively high frequency of nasal carriers of Methicillin-resistant Staphylococcus aureus among the apparently healthy residents of the studied area and the advent of multidrug resistance among these isolates. Our study also supports previous findings on male-gender and low educational background as risk factors of S. aureus carriage. The need for rational chemotherapy, routine detection and regular surveillance of Methicillin-resistant Staphylococcus to limit its spread and reduce treatment failures is important.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Methicillin-resistant Staphylococcus aureus (MRSA) has emerged as an important pathogen in human medicine. MRSA was first reported in 1961, soon after the introduction of methicillin into human medicine to treat penicillin-resistant staphylococci in nosocomial infections (Conly and Johnston 2003; Lakhundi and Zhang 2018). Methicillin resistance is of great importance because it is conferred by the presence of the mecA gene that resides on a staphylococcal chromosomal cassette (SCC), which encodes for the production of an altered penicillin-binding protein (PBP; PBP2a or PBP2) that has a reduced affinity for all beta-lactam antimicrobials. In the past five decades, the incidences of both nosocomial and community-acquired S. aureus (CA-MRSA) infections have increased, while antibiotics treatment options are increasingly hampered by the spread of MRSA (Davis et al. 2007; Yarovoy et al. 2019). MRSA are resistant to a broad range of antimicrobials and are also frequently resistant to most of the commonly used antimicrobial agents such as aminoglycosides, macrolides, chloramphenicol, tetracycline, and fluoroquinolones which make MRSA particularly difficult to treat.
From the systematic review carried out by Abubakar and Sulaiman (2018), the prevalence of MRSA infection in Nigeria was about 50%. Poor infection control, inappropriate use of antibiotics and poor implementation of the developed National action plan for antimicrobial resistance could explain the rising trends of MRSA in Nigeria (FMoH 2017). However, efforts are currently being made to implement such interventions in Nigeria. The emergence of CA-MRSA has changed the epidemiology of S. aureus as reported by several studies (Iwao et al. 2017; Oliveira et al. 2018; Wang et al. 2016). Many studies have characterized S. aureus and MRSA isolates from individuals at some selected communities and hospitals but there is a paucity of information on the distribution of staphylococcal nasal carriage and MRSA among a group of communities in southwestern Nigeria particularly, in Ibadan. Therefore, we sought to determine the distribution, risk factors, and the antibiotic resistance patterns of MRSA to commonly prescribed antibiotics in these localities. This would be useful in choosing empirical therapy for the treatment of CA-MRSA infections and the enactment of infection control guidelines.
2 Methods
2.1 Ethical Approval
Ethical approval was sought from the Health Research Ethics Committee (HREC) of the Institute of Public Health, Obafemi Awolowo University, Ile-Ife, Nigeria. We obtained informed consent from all participants or their legal guardians and confidentiality of all participants and premise data was strictly maintained.
2.2 Study Population and Collection of Samples
This was a cross-sectional, multicenter study using a proportionate stratified random sampling technique. Ibadan city is the state capital of Oyo-State with over 3.7 million inhabitants. The participants were recruited from the 11 local government areas (LGAs) in Ibadan, Nigeria for a period of 9 months (Fig. 1). Nasal specimens were collected by streaking both the anterior nares of each participant using sterile swabs (Copan Diagnostics, Corona, CA, USA) moisture with sterile normal saline. The samples were transported to the laboratory aseptically within 2 h of collection.
Map indicating the location of the study area (Olayinka 2021)
2.3 Isolation and Identification of Staphylococcus aureus
All nasal swabs were inoculated onto Mannitol Salt Agar and Blood Agar (Oxoid Ltd., Hampshire, England) aseptically and incubated aerobically at 37 °C for 24–48 h. Discrete colonies of Staphylococcus aureus were sub-cultured on Nutrient agar (Oxoid Ltd., Basingstoke, Hampshire, England) plates incubated aerobically at 37 °C for 24 h to obtain pure culture and for further analyses. Each isolate was identified using Gram-staining, Cowan & Steel method of bacteria identification, Microbact™ 12S identification kit and confirmed via 16SrRNA gene detection.
2.4 Antimicrobial Sensitivity Testing
The susceptibility of recovered S. aureus isolates to various antibiotics were determined according to Kirby-Bauer disc diffusion technique and the (Clinical and Laboratory Standards Institute 2021) guideline was used to interpret the result. The tested antibiotics include Tetracycline (30 μg), Gentamicin (10 μg), Clindamycin (2 μg), Erythromycin (15 μg), Oxacillin (1 μg), Ceftaroline (30 μg), Co-trimoxazole (25 μg), Linezolid (30 μg), Vancomycin (30 μg), Cefoxitin (30 μg), and Ceftriaxone (30 μg) (Oxoid Ltd., Basingstoke, United Kingdom). We used cefoxitin discs (30 μg) (Oxoid Ltd., Basingstoke, United Kingdom) to phenotypically screen for methicillin-resistance). S. aureus ATCC 25923 was used for quality control.
2.5 Genomic DNA Isolation and Detection of mecA and nuc Genes
Promega (Madison, USA) genomic DNA extraction kit was according to the manufacturers’ instructions using aseptic precautions. Polymerase Chain Reaction (PCR) assay was carried out for mecA (for detection of methicillin resistance) and nuc gene (for detection of S. aureus). The primer sequenced were as follows mec-A1 (5′- AAA ATC GAT GGT AAA GGT TGC C-3′), mec-A2 (5′- AGT TCT GCA GTA CCG GAT TTG C- 3′) and nuc-A1 (5′- GCG ATT GAT GGT GAT ACG GTT-3′), nuc-A2 (5′- AGC CAA GCC TTG AAC GAA CTA AAGC- 3′ (David et al. 2010).
2.5.1 Questionnaire Design
We developed and administered a structured questionnaire. Three independent reviewers were selected to validate the questionnaire; to assess the content validity, clarity, ease of response, scope, and face validity of the questions, We also obtained the participants’ demographic characteristics (gender, education, and age group) and information on risk factors (recent antibiotics use, handwashing frequency, educational background of participants, etc.) for predisposition to colonization (Supplementary file 1).
2.5.2 Statistical Analysis
The data was summarized using Microsoft excel 2016 and subjected to further statistical analysis using Chi-Square and Fisher’s Exact Probability Test in Epi-Info V.7.0 (CDC, Atlanta, USA). Inferences were made based on computed Prevalence ratios, their 95% confidence intervals and p-values. The level of significance was set at p ˂ 0.05.
3 Results
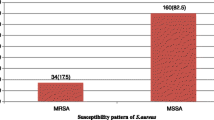
Seven hundred (700) participants were included in this study from their respective LGAs; 56% (392/700) males and 44% (308/700) female participants while the age range (30–40 years) had the highest frequency of participants 20.1% (141/700). Most of the participants (62.6%, n = 438/700) had no formal/primary education (Table 1). The prevalence of S. aureus nasal carriage and MRSA in this study was 31.9% (223/700) and 9.43% (66/700) respectively. MRSA was well distributed in all the eleven (11) LGAs in the study area with the highest prevalence in Egbeda LGA (28.6%, n = 18/63). Results showed that methicillin-resistant S. aureus is significantly associated with participants that reside in Ibadan North LGA (FE = 1.8962, P = .028) and Egbeda LGA (FE = 2.7222, P = .006) (Table 2).
The susceptibility pattern of nasal S. aureus to different antibiotics is presented in Table 3. Most of the MRSA were resistant to other antibiotics such as Oxacillin (96.9%), clindamycin (62.1%), Co-trimoxazole (59.1%) as revealed in Fig. 2. This study showed that nasal Staphylococcus aureus are highly multidrug-resistant as 54% were resistant to two different classes of antibiotics while 24% were resistant to three or more different classes of antibiotics. All the 66 (100%) MRSA isolates harboured the mecA gene (Fig. 3). In addition, 18 (27.3%) of the recovered MRSA possess the nuc gene (Fig. 4).
PCR amplification of MecA gene (533 bp), 16-10-2020
Lane M: DNA molecular size marker (100 bp ladder), Lane 1: ATCC 33591 (positive control), Lane 2: Isolate 14; Lane 3: Isolate 28; Lane 4: Isolate 35; Lane 5: Isolate 38; Lane 6: Isolate 48; Lane 7: Isolate 55; Lane 8: Isolate 62; Lane 9: Isolate 69; Lane 10: Isolate 71.
The result of this study showed that participants between 40–50 years old (34.5%, ϰ2 = 9.842, P = .002) and the male participants (35.3%, ϰ2 = 8.849, P = .003) were more likely to be carriers of MRSA (Table 4). Some of the factors that are associated with MRSA infection were tested in this study; the participants that visited the hospital recently (39.7%, ϰ2 = 8.693, P = .003), used antibiotics recently (61.7%, ϰ2 = 7.556, P = .006) and those with low educational background (3.9%, ϰ2 = 36.817, P ˂ .001) are more likely to be carriers of MRSA as presented in Table 4.
4 Discussion
The epidemiological knowledge of Methicillin-resistant Staphylococcus aureus (MRSA) infection is very important for appropriate decision-making in the treatment of infections. Since at least 1978 when we have the first reported case of MRSA in Africa (Scragg and Appelbaum 1978), we have been attending to an increase in the number of infection episodes in healthy individuals (both adults and children) in the community (Abubakar and Sulaiman 2018). According to the review done by Abdulgader et al. (2015), only fifteen (15) out of the fifty-four (54) African countries are with reports on the molecular epidemiology of MRSA. Additionally, despite the global challenge on the burden of disease caused by CA-MRSA, in Nigeria, only a few isolated studies describing MRSA with typical community backgrounds were reported (Ghebremedhin et al. 2009). In this study, we report for the first time the prevalence, risk factors and the epidemiology of CA-MRSA infections in Ibadan as well as determine the antibiotic resistance indices of MRSA in different communities in Ibadan, Nigeria.
It has been reported that all beta-lactam antibiotics have poor affinity when penicillin-binding protein (PBP) is altered; it will be difficult to kill such microorganisms when exposed to therapeutic concentration. Methicillin resistance is mediated among Staphylococcus aureus by the penicillin-binding protein encoded by the mecA gene (Ito et al. 2014). The overall prevalence of CA-MRSA in this study was 9.43%. The MRSA prevalence reported in this study is higher than what was reported (4%) by Ajani et al. (2020) among students of a private institution in Ogun-State, Nigeria.
We detected nuc genes of S. aureus in only 27.3% of the MRSA isolates this is probably due to the differences in the nucleotide sequence among the nuc genes caused by some mutation or the absence of nuc gene in some S. aureus strains (Sahebnasagh et al. 2014). The difference between the phenotypic and genotypic detection of S. aureus strains made it clear that, the method of identifying only nuc genes in S. aureus is not sufficient. Therefore, both phenotypic and genotypic methods should be conducted for the identification of S. aureus strain. Methicillin-resistant Staphylococcus aureus isolates were obtained from all the 11 LGAs in Ibadan, Nigeria. Egbeda and Ibadan North LGAs presented the highest MRSA prevalence in this study. Individuals that reside in either of these LGAs (Ibadan North; FE = 1.8962, P = .028 or Egbeda; FE = 2.7222, P = .006) are more likely to carry MRSA than those in other areas. This is because the two LGAs are among the most populated LGAs in Ibadan with shared border and linked roadways (NPC 2006; Olayinka 2021). This result is similar to the previous MRSA prevalence range of 14.3–37% that has been reported in different communities in Nigeria (Akerele et al. 2015; Bale et al. 2019; Egwuatu et al. 2016; Ghebremedhin et al. 2009; Taiwo et al. 2005). This is not surprising as the MRSA prevalence varies greatly with geographical location and studied population (Bell et al. 2002).
Antimicrobial resistance (AMR) is one of the major threats posed by microorganisms in this twenty-first century. Despite the serious efforts employed to control AMR by aggressive infection control methods, MRSA has become one of the most frequent cause of hospital and community-acquired infections globally. MRSA has always been one of the major pathogens that possess the ability to develop resistance to newly developed antimicrobial agents (Joo et al. 2017). In this study, Staphylococcus aureus nasal carriage exhibited resistance to Oxacillin, Co-trimoxazole and Clindamycin but susceptible to Vancomycin, Gentamicin, Tetracycline, Ceftaroline and Linezolid in varying degrees. The MRSA isolates were also resistant to multiple antibiotics. The antimicrobial resistance patterns of the isolates revealed high (80%) multidrug resistance (resistance against at least 2 different classes of antimicrobials) rate among the MRSA isolates. Shariati et al. (2020) reported a global increase in the prevalence of vancomycin resistance in their systemic review of global prevalence and distribution of vancomycin-resistant Staphylococcus aureus. Therefore, the 7.6%prevalence of vancomycin resistance in this study is not surprising although it remains a major global public health challenge. The relatively high resistance prevalence observed in both Ceftaroline and linezolid in this study are comparable with the rate of resistance reported in a neighboring town by Osinupebi et al. (2018). Many factors can be linked to the high resistance of this organism to antibiotics in these communities. Such factors include self-medication, availability and use of antibiotics without prescription, irrational consumption rate of antibiotics, over the counter accessibility without prescription and sales of fake or substandard drugs, unrestricted use of antimicrobials in farm animals (including poultries and fisheries), and transmission of resistant strains between individuals within the community (Akerele et al. 2015; Elimam et al. 2014).
One of the main reason for mecA gene screening by polymerase chain reaction (PCR) technique is to compare the results of antibiotic susceptibility by disc diffusion method with gene analysis results in Staphylococcus aureus isolates. Not all MRSA strains may be detectable with phenotypical methods as some mecA genes may be heterogeneously expressed. In this study, all the MRSA isolates that were cefoxitin resistant were also positive for this gene detection method confirming that they are all methicillin-resistant Staphylococcus aureus. The study showed a strong correlation between genotypic and phenotypic analysis and it is consistent with previous studies that reported a perfect association between the results obtained by the phenotypic antibiotic resistance determination and PCR- based assays (Bale et al. 2019; Ito et al. 2014; Strommenger et al. 2006).
The present study showed a high proportion of CA-MRSA in the male-gender than in female which agrees with previous studies (Abroo et al. 2017; Mehraj et al. 2014; Skramm et al. 2011). The microbial differences between male and female could be due to physiological factors or anatomical differences between genders (Giacomoni et al. 2009). Our findings that males were more likely to carry MRSA is consistent with other studies, which indicate a gender-specific risk factor (Andersen et al. 2013; Assafi et al. 2015; Gorwitz et al. 2008; Graham et al. 2017; Mehraj et al. 2014; Skramm et al. 2011). Some studies have reported the association of MRSA among adults and elderly as revealed in this study (Andersen et al. 2013; Gorwitz et al. 2008). This study also revealed that individuals with certain risk factors (age, male gender, recent use of antibiotics, recent hospital visitation, and level of education) were more likely to be carriers of MRSA (Abroo et al. 2017; Graham et al. 2017).
5 Conclusion
In conclusion, the strongest risk factors of CA-MRSA were male gender, low educational background, recent antibiotics used, and hospital visits. The relatively high prevalence of CA-MRSA in this study is a cause for concern. CA-MRSA showed a high resistance burden and individuals who are harboring these isolates can act as reservoirs; this may negatively influence the treatment of CA-MRSA infections. The continuous surveillance of CA-MRSA is essential to prevent transmission of Staphylococcus aureus from the infected carriers to others also to apply effective therapeutic options for their treatment.
References
Abdulgader SM, Shittu AO, Nicol MP, Kaba M (2015) Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Africa: a systematic review. Front Microbiol 6(April). https://doi.org/10.3389/fmicb.2015.00348
Abroo S, Jazani NH, Sharifi Y (2017) Methicillin-resistant Staphylococcus aureus nasal carriage between healthy students of medical and nonmedical universities. Am J Infect Control 45(7):709–712. https://doi.org/10.1016/j.ajic.2017.02.034
Abubakar U, Sulaiman SAS (2018) Journal of Infection and Public Health Prevalence, trend and antimicrobial susceptibility of methicillin resistant Staphylococcus aureus in Nigeria: a systematic review. J Infect Public Health 11(6):763–770. https://doi.org/10.1016/j.jiph.2018.05.013
Ajani TA, Elikwu CJ, Nwadike V, Babatunde T, Anaedobe CG, Shonekan O, Okangba CC, Omeonu A, Faluyi B, Thompson TE, Ebeigbe E, Eze BG, Ajani MA, Perelade K, Amoran M, Okisor P, Worancha T, Ayoade J, Agbeniga E et al (2020) Nasal carriage of methicillin resistant Staphylococcus aureus among medical students of a private institution in Ilishan-Remo, Ogun State, Nigeria. Afr J Clin Exp Microbiol 21(4):311–317
Akerele JO, Obasuyi O, Omede D (2015) Prevalence of methicillin-resistant Staphylococcus aureus among healthy residents of Ekosodin Community in Benin-City, Nigeria. Trop J Pharm Res 14(August):1495–1499
Andersen PS, Larsen LA, Jr VGF (2013) Risk factors for Staphylococcus aureus nasal colonization in Danish middle-aged and elderly twins. Eur J Clin Microbiol Infect Dis 32:1321–1326. https://doi.org/10.1007/s10096-013-1882-0
Assafi MS, Mohammed RQ, Hussein NR (2015) Nasal carriage rates of Staphylococcus aureus and CA-methicillin resistant Staphylococcus aureus among university students. J Microbiol Res. https://doi.org/10.5923/j.microbiology.20150504.01
Bale MI, Babatunde SK, Adedayo MR, Ajiboye AE, Ajao AT (2019) Characterization of methicillin-resistant Staphylococcus aureus isolates from apparently healthy individuals in Malete, Kwara State, Nigeria. Afr J Clin Exp Microbiol 20(1):17–24
Bell JM, Turnidge JD, Participants SA (2002) High prevalence of oxacillin-resistant Staphylococcus aureus isolates from hospitalized patients in Asia-Pacific and South Africa: results from SENTRY antimicrobial surveillance program, 1998–1999. Antimicrob Agents Chemother 46(3):879–881. https://doi.org/10.1128/AAC.46.3.879
Clinical and Laboratory Standards Institute (CLSI) (2021) Supplement M100: performance standards for antimicrobial susceptibility testing (Kristy LL, Laura M, eds.), 31st ed. Clinical and Laboratory Standards Institute, Wayne,
Conly JM, Johnston BL (2003) The emergence of as a community-acquired pathogen in Canada. Can J Infect Dis 14(5):249–251
David MZ, Daum RS, David MZ, Daum RS (2010) Methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic community-associated methicillin-resistant Staphylococcus aureus: epidemiology and clinical consequences of an emerging epidemic. Clin Microbiol Rev 23(3):616–687. https://doi.org/10.1128/CMR.00081-09
Davis SL, Perri MB, Donabedian SM, Manierski C, Singh A, Vager D, Haque NZ, Speirs K, Muder RR, Hayden MK, Zervos MJ, Icrobiol JCLINM (2007) Epidemiology and outcomes of community-associated methicillin-resistant Staphylococcus aureus infection. J Clin Microbiol 45(6):1705–1711. https://doi.org/10.1128/JCM.02311-06
Egwuatu T, Iwuafor A, Nnachi A, Aghanya I (2016) Time-kill effect of crude extracts of Garcinia kola seeds on methicillin-resistant Staphylococcus aureus from the anterior nares of healthcare workers at a Tertiary Hospital in Nig … time-kill effect of crude extracts of Garcinia kola seeds on methicil. J Adv Med Pharm Sci. https://doi.org/10.9734/JAMPS/2016/27005
Elimam M, Rehan S, Elmekki M, Elhassan M (2014) Emergence of vancomycin resistant and methcillin resistant Staphylococus aureus in patients with different clinical manifestations in Khartoum State. J Am Sci 10(6):106–110
FMoH (2017) National Action Plan for antimicrobial resistance 2017–2022 (Aruna S, ed.; 1st edn). FMoH (Federal Ministries of Health), Abuja, Nigeria
Ghebremedhin B, Olugbosi MO, Raji AM, Layer F, Bakare RA, Ko W (2009) Emergence of a community-associated methicillin-resistant Staphylococcus aureus strain with a unique resistance profile in Southwest Nigeria. J Clin Microbiol 47(9):2975–2980. https://doi.org/10.1128/JCM.00648-09
Giacomoni PU, Mammone T, Teri M (2009) Gender-linked differences in human skin. J Dermatol Sci 55:144–149. https://doi.org/10.1016/j.jdermsci.2009.06.001
Gorwitz RJ, Kruszon-moran D, Mcallister SK, Mcquillan G, Mcdougal LK, Fosheim GE, Jensen BJ, Killgore G, Tenover FC, Kuehnert MJ (2008) Changes in the prevalence of nasal colonization with Staphylococcus aureus in the United States, 2001–2004. JID 197:2001–2004. https://doi.org/10.1086/533494
Graham PL, Lin SX, Larson E (2017) A U.S. population-based survey of Staphylococcus aureus colonization. Ann Intern Med 2002(1):2001–2002
Ito T, Kuwahara-Arai K, Katayama Y, YUehara Y, Han X, Kondo Y, Hiramatsu K (2014) Staphylococcal cassette chromosome mec (SCC mec) analysis of MRSA. Methods Mol Biol 1085:131–148. https://doi.org/10.1007/978-1-62703-664-1
Iwao Y, Wakayama M, Inomata N, Takano T, Yamamoto T (2017) Genomic comparison between Staphylococcus aureus GN strains clinically isolated from a familial infection case: IS 1272 transposition through a novel inverted repeat-replacing mechanism. PLoS One 12(11):1–29
Joo E, Chung DR, Kim SH, Baek JY, Lee NY, Cho SY, Ha YE, Kang C, Peck KR, Song J (2017) Emergence of community-genotype methicillin-resistant Staphylococcus aureus in Korean hospitals: clinical characteristics of nosocomial infections by community-genotype strain. Infecti Chemother 49(2):109–116
Lakhundi S, Zhang K (2018) Methicillin-resistant Staphylococcus aureus: molecular characterization, evolution, and epidemiology. Clin Microbiol Rev 31(4):1–103
Mehraj J, Akmatov MK, Strompl J, Gatzemeier A, Layer F, Werner G (2014) Staphylococcus aureus nasal carriage in a random sample of non-hospitalized adult population in northern Germany. PLoS One 9(9). https://doi.org/10.1371/journal.pone.0107937
NPC (2006) National Population Commission, 2006 – Nigeria Data Portal. Nigeria Data Portal. https://nigeria.opendataforafrica.org/ifpbxbd/state-population-2006
Olayinka AA (2021) Map of Ibadan Metropolis. https://www.google.com.ng/maps/@7.4051008,3.8178917,11z/data=!3m1!4b1!4m2!6m1!1s11vkSp3Yg9nAHPKsvLLSHVODYuRKs3ysi?hl=en
Oliveira D, Borges A, Simões M (2018) Staphylococcus aureus toxins and their molecular activity in infectious diseases. Toxins. https://doi.org/10.3390/toxins10060252
Osinupebi OA, Osiyemi JA, Akinduti PA (2018) Prevalence of methicillin-resistant Staphylococcus aureus in Abeokuta, Nigeria. South Asian J Res Microbiol 1(1):1–8. https://doi.org/10.9734/SAJRM/2018/v1i1718
Sahebnasagh R, Saderi H, Owlia P (2014) The prevalence of resistance to methicillin in Staphylococcus aureus strains isolated from patients by PCR method for detection of mecA and nuc genes. Iran J Public Health 43(1):84–92
Scragg JN, Appelbaum IC (1978) The spectrum of infection and sensitivity of organisms isolated African and Indian children in a Durban hospital. Trans R Soc Trop Med Hyg 72(4):325–328
Shariati A, Dadashi M, Moghadam MT, Van Belkum A (2020) Global prevalence and distribution of vancomycin resistant, vancomycin intermediate and heterogeneously vancomycin intermediate Staphylococcus aureus clinical isolates: a systematic review and meta – analysis. Sci Rep:1–16. https://doi.org/10.1038/s41598-020-69058-z
Skramm I, Moen AE, Bukholm G (2011) Nasal carriage of Staphylococcus aureus: frequency and molecular diversity in a randomly sampled Norwegian community population. APMIS 119(4):522–528. https://doi.org/10.1111/j.1600-0463.2011.02758.x
Strommenger B, Kettlitz C, Weniger T, Harmsen D, Friedrich AW, Witte W (2006) Assignment of Staphylococcus isolates to groups by spa typing, SmaI macrorestriction analysis, and multilocus sequence typing. J Clin Microbiol 44(7):2533–2540. https://doi.org/10.1128/JCM.00420-06
Taiwo SS, Bamidele M, Omonigbehin EA, Akinsinde KA, Smith SI, Onile BA, Olowe AO (2005) Molecular epidemiology of methicillin-resistant Staphylococcus aureus in Ilorin, Nigeria. West Afr J Med 24:100–106. https://doi.org/10.4314/wajm.v24i2.28176
Wang X, Li X, Liu W, Huang W, Fu Q, Li M (2016) Molecular characteristic and virulence gene profiles of methicillin-resistant Staphylococcus aureus isolates from Pediatric patients in Shanghai, China. Front Microbiol 7(November). https://doi.org/10.3389/fmicb.2016.01818
Yarovoy JY, Monte AA, Knepper BC, Young HL (2019) Epidemiology of community-onset Staphylococcus aureus Bacteremia. West J Emerg Med 20(May):438–442. https://doi.org/10.5811/westjem.2019.2.41939
Competing Interests
Authors have declared that no competing interests exist.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
1 Supplementary Electronic Material (S)
Supplementary file 1
(DOCX 18 kb)
Rights and permissions
Copyright information
© 2021 The Author(s), under exclusive license to Springer Nature Switzerland AG
About this chapter
Cite this chapter
Oginni, I.O., Olayinka, A.A. (2021). Distribution and Antibiotics Resistance Pattern of Community-Acquired Methicillin-Resistance Staphylococcus aureus in Southwestern Nigeria. In: Donelli, G. (eds) Advances in Microbiology, Infectious Diseases and Public Health. Advances in Experimental Medicine and Biology(), vol 1369. Springer, Cham. https://doi.org/10.1007/5584_2021_658
Download citation
DOI: https://doi.org/10.1007/5584_2021_658
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-031-01994-4
Online ISBN: 978-3-031-01995-1
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)