Abstract
Chronic rhinosinusitis (CRS) is the most common illness among chronic disorders that remains poorly understood from a pathogenic standpoint and has a significant impact on patient quality of life, as well as healthcare costs. Despite being widespread, little is known about the etiology of the CRS. Recent evidence, showing the presence of biofilms within the paranasal sinuses, suggests a role for biofilm in the pathogenesis. To elucidate the role of biofilm in the pathogenesis of CRS, we assessed the presence of biofilm at the infection site and the ability of the aerobic flora isolated from CRS patients to form biofilm in vitro. For selected bacterial strains the susceptibility profiles to antibiotics in biofilm condition was also evaluated.
Staphylococci represented the majority of the isolates obtained from the infection site, with S. epidermidis being the most frequently isolated species. Other isolates were represented by Enterobacteriaceae or by species present in the oral flora. Confocal laser scanning microscopy (CLSM) of the mucosal biopsies taken from patients with CRS revealed the presence of biofilm in the majority of the samples. Strains isolated from the specific infection site of the CRS patients were able to form biofilm in vitro at moderate or high levels, when tested in optimized conditions. No biofilm was observed by CLSM in the biopsies from control patients, although the same biopsies were positive for staphylococci in microbiological culture analysis. Drug-susceptibility tests demonstrated that the susceptibility profile of planktonic bacteria differs from that of sessile bacteria in biofilms.
Access provided by CONRICYT-eBooks. Download chapter PDF
Similar content being viewed by others
Keywords
- Adherent bacteria
- Antibiotic resistance
- Sinus infection
- Isothermal microcalorimetry
- Confocal fluorescence microscopy
1 Introduction
Rhinosinusitis (RS) comprises a group of disorders characterized by inflammation of the mucosa of the nose and the paranasal sinuses (Lanza and Kennedy 1997). According to the duration of the symptoms, RSs are sub-divided into acute forms, in which symptoms last up to 12 weeks followed by complete resolution, and chronic forms in which symptoms persist beyond 12 weeks. Chronic rhinosinusitis (CRS), whose prevalence is estimated to be approximately 10% of the population in the Western world, is considered a multifactorial disease in which both host-related (e.g. anatomical features, immunological status, ciliary dysfunction, associated co-morbidities) and non-host-related factors (e.g. microbial agents, allergens) may play a role (Fokkens et al. 2012; Lam et al. 2015). The etiology and pathogenesis of CRS remain an area of active research. In this respect, it is still debated whether bacterial infections, in the form of biofilms, might be involved in the pathogenesis of CRS contributing to the inflammation, persistence, and recurrent exacerbations of the disease (Lam et al. 2015). Bacterial biofilms are communities of sessile bacteria embedded in a protective extracellular polymeric substance (EPS) they have produced, and exhibiting a phenotype markedly different from the corresponding planktonic cells concerning the growth rate and gene expression (Donlan and Costerton 2002). It is becoming increasingly evident that biofilms are involved in a large proportion of infections of the human body and may form on host tissues/mucosa or on the surface of a variety of medical devices including central vascular or urinary catheters, endotracheal tubes, prosthetic cardiac valves, orthopedic or dental implants (Lebeaux et al. 2014). Hallmark of biofilm-associated infections is their dramatically reduced susceptibility to antimicrobial treatments that is considered a multifactorial process (Lebeaux et al. 2014). In addition to the classical mechanisms of resistance at the single cell level (e.g. activation of efflux pumps, production of enzymes that destroy antibiotics, mutations that alter antibiotic target sites), multiple biofilm-specific mechanisms operate simultaneously and make it difficult to completely kill cells in a biofilm, especially those situated in the deeper layers (Penesyan et al. 2015; Sun et al. 2013). Consequently, biofilm-associated infections usually do not resolve easily, requiring the mechanical or surgical removal of the biofilm-infected tissue or colonized implant.
In addition to the tolerance to antimicrobial treatments, biofilms also exhibit a high capacity to resist the clearance by host innate and adaptive immune responses (Lebeaux et al. 2014). Both of these factors play a major role in treatment failure and persistence of biofilm-associated infections.
Biofilm-associated infections are not only difficult to treat, but also to diagnose by routine, culture-based, microbiological techniques (Trampuz et al. 2007). Indeed, methods traditionally used in clinical microbiology laboratories are optimized for detecting and culturing infectious microorganisms growing in planktonic (free floating) forms and to test their susceptibility under planktonic growth conditions (Hoiby et al. 2015). Due to the marked ability of biofilm cells to adhere to the substrate and/or to each other via the EPS, biofilm-embedded microorganisms might be much more difficult to recover from clinical samples than planktonic ones. Hence, the culture of biofilm cells from infected tissues is often negative, despite the presence of large aggregates of bacteria surrounded by an extracellular matrix can be demonstrated on tissues or medical devices by microscopic techniques (Hoiby et al. 2015; Batoni et al. 2016). Failure to isolate microbial strain(s) possibly involved in the pathogenic process, in turn, further affects the outcome of the antimicrobial therapy, in that the clinicians lack a rational basis for antibiotic selection (Hall-Stoodley et al. 2012).
In order to gain further insights on the possible role of biofilm-infections in CRS and diagnostic procedures suitable to aid laboratory diagnosis of such infections (e.g. type and mode of sampling, sample processing procedures, culture conditions etc.), the purposes of the present study were: (i) evaluate the presence of bacterial aggregates resembling biofilm structures in biopsies from CRS patients selected according to stringent clinical criteria (Benninger et al. 2003; Rosenfeld et al. 2015); (ii) isolate and identify aerobic bacterial strains from biopsies and other clinical samples collected from diseased and non-diseased areas by microbiological diagnostic techniques; (iii) assess the in vitro biofilm-forming ability of bacteria isolated from infection sites in different experimental conditions; (iv) compare the antibiotic susceptibility profile of the isolated strains in planktonic form versus that obtained in biofilm state.
The consistent demonstration of structured sessile bacteria on biopsies from CRS patients, and their marked ability to form biofilms in vitro, when tested in optimized experimental conditions, corroborates the hypothesis that biofilms may play a role in the pathogenesis/chronicity of the disease. The results obtained also suggest that modifications of routine microbiological procedures might be necessary to increase the rate of culture positivity. Finally, the choice of an eventual antimicrobial therapy to treat/prevent CRS could benefit from evaluation of the antibiotic sensitivity profiles of sessile bacteria.
2 Materials and Methods
2.1 Study Group and Sample Collection
The study group consisted of 12 patients with CRS, undergoing functional endoscopic sinus surgery (FESS). In addition, 5 patients undergoing septoplasty surgery for a surgical correction of a deviated nasal septum and reduction of the volume of the inferior turbinate with no clinical or radiological evidence of sinus disease or allergic rhinitis were selected as negative controls (1C-5C, Table S1). CRS patients were recruited after an accurate medical examination, according to the guidelines of the Rhinosinusitis task force (Benninger et al. 2003; Rosenfeld et al. 2015). In particular, medical examination was aimed at verifying the inflammation status of the nose and of the paranasal sinuses and its duration (minimum 12 weeks), and the presence of at least two of the following symptoms: nasal blockage/obstruction/congestion, nasal discharge (anterior/posterior nasal drip), facial pain/pressure, reduction or loss of smell, endoscopic signs of the disease. Clinical diagnosis of CRS was confirmed by using computerized tomography (CT) allowing a differential diagnosis with other diseases with similar symptoms such as upper respiratory tract infections or allergic rhinitis (Desrosiers et al. 2011).
Exclusion criteria included pregnancy, immunodeficiency, and impairment of muco-ciliary function. Preoperative data collection included symptom scores, allergy status, paranasal sinus computed tomography (CT) scores, past medical history, smoking status, and nasal endoscopy findings. No patients had taken antibiotics, antifungals, or steroids in the 4 weeks prior to surgery.
Samples were collected at the Otorhinolaryngology Unit of the Department of Surgery and Medicine, at the Pisa University Hospital, Pisa, Italy.
Nasal mucosa biopsies were harvested from CRS and control patients at the time of FESS and nasal septoplasty, respectively, and were placed in a sterile container. One to five pieces (size: ≤ 5 × 5 mm) were taken from each patient for confocal laser scanning microscopy (CLSM) and microbiological examination. Only the biopsy of the patient #9, was analysed by microbiological examination and not by CLSM due to its small size. In addition, sinus swabs from the site of the surgery and/or sinus aspirates were collected for bacterial and fungal cultures/identifications. Extreme attention was paid to avoid contamination with normal flora during sample collection. For some patients a nasal swab was also collected to evaluate the nose normal flora.
The Human Ethics Committee of the Pisa University Hospital approved the study and all patients provided their informed consent to participate in the study.
2.2 Detection of Biofilm-Like Structures in Biopsies from CRS and Control Patients by CLSM
The presence of bacterial aggregates resembling bacterial biofilms on biopsy samples (processed as described in paragraph 2.3) was evaluated by CLSM analysis as previously described (Foreman et al. 2009, 2010; Psaltis et al. 2007). Briefly, fresh specimens were immersed in 1 ml of sterile MilliQ water (Millipore), within 3 h from collection and added with 1.5 μl of Syto9 (Thermo Fisher Scientific). After incubation in the dark at room temperature for 15 min, each biopsy was rinsed in sterile MilliQ water to remove the excess of stain and placed on glass bottom dishes (Willco Wells) for microscopy imaging. In the case it was not possible to perform the analysis immediately, biopsy samples were fixed with 4% PFA for 4 h at 4 °C, washed three times with PBS, and stored at 4 °C in PBS added with 0.05% (w/v) NaN3 to prevent microbial contamination, until staining and imaging. Stained specimens were observed under TCS SP5 II (Leica) confocal microscope (interfaced with a 488 nm Argon laser), using a 63 × 1.25 NA water immersion objective. For each biopsy the entire tissue surface and depth were scanned during three independent observations for a total of 45 min. 10 μm Axial stacks in the Z plane, with a slice thickness of 1 μm, were taken through representative areas of biofilm. The analysis was not performed blind.
2.3 Isolation, Identification and Planktonic Drug Susceptibility Testing of Clinical Isolates
Biopsies, sinus swabs, and/or sinus aspirates were transferred within 1–3 h from collection to the Microbiology Unit laboratory of Pisa University Hospital, and processed as follows: biopsies were washed three times in PBS to remove planktonic bacteria and divided into two pieces with a scalpel under sterile conditions. One of the pieces was then processed for staining and imaging as described in paragraph 2.2. The other piece was homogenised in 1 ml of PBS using a bead beater instrument (Stomaker), and vigorously vortexed for 1 min at room temperature. When multiple biopsies from the same patient were collected, they were individually processed for CLMS imaging, while they were pooled and homogenized all together for microbiological analyses. To this aim, 0.1 ml aliquots of the homogenates were inoculated directly onto blood agar, chocolate agar, mannitol salt agar, MacConkey agar, and Sabouraud agar, and cultured for 24–48 h at 37 °C in aerobiosis. The remaining homogenised samples were inoculated in 5 ml of brain heart infusion broth (BHIB, Becton Dickinson). After 24 h incubation at 37 °C, turbid broth samples were sub-cultured onto the five agar plates listed above. Selected colonies were identified using the matrix-assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF) system (Bruker) using procedures suggested by the manufacturer. Swabs from nasal mucosa or from diseased sites were streaked directly onto the agar plates and cultured as described above. Sinus aspirates were processed as biopsies omitting the homogenization step.
For all isolated bacterial strains routine antibiotic susceptibility tests were performed by semiautomatic systems (Vitek II, bioMérieux) and/or Sensititre (Thermo Scientific).
2.4 Cultivation and Storage of Bacterial Strains
A single colony of bacterial isolates and standard laboratory strains (Staphylococcus epidermidis ATCC35984 and S. aureus ATCC43300) used as controls for biofilm production, were inoculated into 5 ml of Tryptone Soy Broth (TSB) (Oxoid,). Haemophilus parainfluenzae and H. paraphrophilus strains were cultivated in Haemophilus Test Medium Broth (Becton Dickinson). Actinomyces odontolyticus, Streptococcus spp., Corynebacterium spp., and Moraxella spp. strains were cultivated in BHIB. After overnight incubation at 37 °C in shaking conditions, an aliquot of the cultures was re-inoculated in the same medium and the cultures were allowed to grow to an OD600 of 1. Each culture was divided into 0.25 ml aliquots, labelled and stored at −80 °C until use.
2.5 Stock Dilutions and Storage of Antibiotics
The antibiotics vancomycin hydrochloride, amoxicillin, amikacin, levofloxacin, doxycycline (all purchased from Sigma-Aldrich) were diluted in MilliQ sterile water (Millipore) to obtain stock solutions of 100 mg/ml. Erythromycin (Sigma-Aldrich) was diluted in pure ethanol (50 mg/ml) while rifampicin and daptomycin were diluted in DMSO (100 mg/ml). All the stock solutions were divided in aliquots and kept frozen at −80 °C or at 4 °C until use, as suggested by the producers.
2.6 Microtiter Plate Biofilm Formation Assay and Crystal Violet (CV) Staining
In a first set of experiments, overnight cultures of selected clinical isolates were diluted to 0.05 OD600 in TSB or Mueller Hinton Broth 2 cation adjusted (MHCA) (Sigma-Aldrich) added with 1% (w/v) glucose (Sigma-Aldrich) (MHCA/Glu) or 1% (w/v) sucrose (Sigma-Aldrich) (MHCA/Suc).
For selected species/strains additional biofilm formation assays were performed in MHCA or MHCA/Gluc added with 1% (w/v) NaCl, or in MHCA/Glu added with 1% (v/v) pooled heat inactivated human plasma collected from healthy donors.
Two hundred μl of each diluted bacterial suspension was dispensed into flat-bottom polystyrene 96-well plates (Corning Costar). Wells with medium alone were set up as negative controls. Plates were incubated at 37 °C without shaking for 24 h. After incubation, wells were washed three times with PBS, air-dried for 15 min at room temperature, and CV (Sigma-Aldrich) was added to a final concentration of 0.5% (w/v). Following incubation for 15 min at room temperature, wells were rinsed with PBS until no blue was visible in the washing solution, air-dried and CV was extracted by incubation for 15 min with pure ethanol (Sigma-Aldrich) at room temperature. The OD was measured at a wavelength of 595 nm with the INFINITE F50 (TECAN) absorbance microplate reader. The assays were performed in triplicate, and the results expressed as mean OD595 ± the standard deviation (SD). Based on the values of OD595 obtained, all strains were classified for their ability to form biofilms into four categories (no, weak, moderate and strong producers) according to Stepanovic and co-workers (Stepanovic et al. 2007).
2.7 CLSM and Image Analysis of In Vitro Formed Biofilms
CLSM and image analysis were used to analyze the architecture and extracellular matrix components of biofilms formed in vitro by staphylococcal clinical isolates, as previously described (Brancatisano et al. 2014; Maisetta et al. 2016). To this aim, 300 μl of bacterial suspensions, diluted as described above for microtiter plate biofilm formation assay, was dispensed into an 8-well μ-Slide (Ibidi) and statically incubated at 37 °C for 24 h. Following incubation, medium was carefully removed from the wells, biofilms were washed three times with 300 μl MilliQ water, and stained with 300 μl Syto9 (1.5 nM) for observation with CLSM, as suggested by the producer (Thermo Fisher Scientific). For the characterization of the staphylococcal biofilm extracellular matrix, biofilms were incubated in the dark for 20 min with 300 μl of undiluted FilmTracer™ SYPRO® Ruby biofilm matrix (Life Technologies) containing 5.0 μg/ml Wheat Germ Agglutinin, Oregon Green® 488 Conjugate (WGA488) (Life Technologies). After staining, biofilms were washed three times with pure water and examined under a Leica CLSM, as described above. An argon laser was used to excite the fluorophores at wavelengths of 458 nm for SYPRO Ruby and 488 nm for WGA and syto-9. The following collection ranges were adopted: 500–540 nm (WGA488 and Syto9), and 605–650 (SYPRO Ruby). In a typical two-channel experiment, images were collected in sequential mode to eliminate emission cross-talk or bleed-through between the various dyes. Two independent experiments were performed and 10 images for each sample were collected for biofilm formation and matrix component analysis.
2.8 Colony Morphology on Congo Red Agar
Staphylococcal strains isolated from CRS patients were tested for polysaccharide intercellular adhesin (PIA) production by the Congo red agar (CRA) method as described by Freeman and co-workers (1989) with minor modifications. To this aim, bacterial strains were plated on CRA (Mueller–Hinton broth 21 g l − 1, 1% bacteriological agar (Oxoid), glucose 50 g l − 1, and Congo red 0.8 g l − 1 [Sigma-Aldrich]). Plates were incubated for 24 h at 37 °C and for an additional 24 h at room temperature. Following incubation, the color and the morphology of the colonies were macroscopically examined. Black/gray colonies with a dry crystalline morphology allowed the recognition of PIA-producing strains, while red/pink, and smooth colonies were considered to be PIA negative.
2.9 Colorimetric Antibiotic Susceptibility Testing of Bacterial Biofilms by Alamar Blue (AB)
Biofilm susceptibility testing was assessed by microplate AB assay as described by Pettit and co-workers (2005). Briefly, biofilms were let to form for 24 h in MHCA/Glu medium or, in the case of S. aureus, in MHCA/Glu added 1% with human inactivated plasma as described above. Biofilms were washed three times with PBS and 200 μl of two-fold serial dilutions of antibiotics in fresh MHCA/Glu medium were dispensed into each well. Biofilms, exposed to drugs for 24 h at 37 °C, were washed three times with PBS and 5 μl of the oxidation reduction indicator AB was added to each well. Then, plates were shaken gently and incubated for 1 h at 37 °C. Color changes in the wells, from blue to pink or purplish, were visually recorded at the end of the incubation to assess the minimum biofilm inhibiting concentrations (MBICs), defined as the lowest drug concentration resulting in ≤50% reduction of AB and a purplish well 60 min after the addition of AB (Pettit et al. 2005).
2.10 Antibiotic Susceptibility Testing of S. aureus and P. mirabilis Biofilms by Isothermal Microcalorimetry Assay
Biofilm antibiotic susceptibility testing was performed by isothermal microcalorimetry (IMC) according to the procedure described by Oliva and co-workers (2014). Briefly, bacterial biofilms were formed on porous glass beads having a diameter 2 to 4 mm, porosity 0.2 m2/g and pore size 60 μm (Siran carrier; SiKUG 023/02/300/A; Schott Schleiffer AG, Muttenz, Switzerland). To this aim, beads were incubated for 24 h at 37 °C in MHCA with 3–5 colonies of S. aureus or 1 colony of P. mirabilis (≈1x10^8 CFU). Then, beads were washed and incubated an additional 24 h at 37 °C in the presence of two-fold serially diluted antibiotics at concentrations up to 1024 μg/ml. Following a second wash to remove non-attached bacteria and antibiotics, each bead was transferred into a different ampoule containing fresh medium without any antibiotics. Finally, all the ampoules were air-tightly sealed and inserted into the microcalorimeter to quantify the heat flow produced by viable bacteria. An isothermal microcalorimeter (TAM III, TA Instruments, Newcastle, USA), equipped with 48 independent channels and a detection limit of heat production of 0.2 μW was used. Growth media with biofilm on beads but without antibiotic pre-treatment were used as positive controls, while growth medium with sterile beads served as negative control. Experiments were performed in triplicates for each strain and the heat flow was recorded for about 48 h.
The minimum biofilm eradicating concentration (MBEC) was defined as the lowest concentration that eradicated biofilm and led to an absence of re-growth after 48 h of incubation in the microcalorimeter (Oliva et al. 2014).
2.11 Data Analysis
OD595 data of CV experiments were collected by spectrophotometer, analysed using Magellan software (TECAN), and exported/plotted using Microsoft Excel program.
Digital images of the CLSM optical section were collected using LAS-AF software (Leica, Heidelberg, Germany) and processed by ImageJ software (NIH Imagej; http://rsbweb.nih.gov/ij/). Microcalorimetry data were analyzed using the manufacturer’s software (TAM Assistant, TA Instruments, New Castle, DE). Figures were plotted using GraphPad Prism 6.01 (GraphPad Software, La Jolla, CA, USA).
2.12 Statistical Analyses
The statistical significance of the data was determined by Fisher’s exact test. A two-tailed P value of <0.05 was considered statistically significant.
3 Results and Discussion
3.1 Study Group and Demographic Data
A total of 17 patients were recruited for this study. Among these, 12 were patients with CRS undergoing FESS, and 5 were negative controls (patients undergoing septoplasty surgery).
The CRS patients’ group consisted of 6 females and 6 males, (median = 53, range 28–78 yrs), The wide age-range of the study group is in agreement with data from the literature reporting that although CRS affects persons of all age groups, its incidence increases with age, peaking at 50 years or greater (Peters et al. 2014). The control group was made of 3 females and 2 males (median = 40, range 22–55 yrs). From a clinical point of view, the CRS patients included 3 subjects with nasal polyposis and 9 patients without polyposis. The demographic data, the clinical diagnosis, and the types of samples collected for CLSM analysis and microbiological examination from each patient and control are reported in supplementary information (Table S1).
3.2 Detection of Bacterial Biofilms in Biopsies from CRS and Control Patients by Syto9 Staining and CLSM Analysis
Biopsies from sinus mucosa of CRS and control patients were analyzed for the presence of biofilms by CLSM following fluorescence staining. Analysis of the biopsies was performed using a previously described, easy, and rapid protocol for the search of bacterial aggregates in tissue samples with minor modifications (Foreman et al. 2009; Foreman et al. 2010; Psaltis et al. 2007). The used probe specifically stains DNA of living and dead cells, including epithelial cells and bacterial cells, as well as the biofilm exopolysaccharide matrix, which contains bacterial DNA. Discrimination of human cells from bacterial cells is based on the different size and shape of the stained cells and, in some cases, to differences in fluorescence intensity between bacterial and host cells, since bacteria stain brighter as compared to eukaryotic cells.
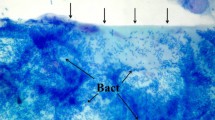
Seven out of 11 CRS patients analyzed by CLSM had biofilms detectable in their biopsies. Figure 1 shows representative magnified images of biofilm-like structures detected in the biopsy samples of patient #1, #4, #5, #6 and #12. The characteristic biofilm morphology with brightly fluorescent microorganisms, whose size is compatible with that of bacterial cells (ranging from 0.5 to 3 μm in most of the cases or longer) is clearly visible. In 3 out of 7 CRS patients whose biopsies were found biofilm-positive, only cocci or filament shaped bacteria were detected. In contrast, CLSM images of biopsies from patients #4 (Fig. 1B), #5 (Fig. 1C–D) and #12 (Fig. 1F) revealed the presence of bacterial cells with two different shapes (cocci and filaments), consistent with the isolation from these biopsies of Staphylococcus lugdunensis/Citrobacter koseri (#4), S. aureus/Proteus mirabilis (#5), and S. epidermidis, S. capitis/Haemophilus parainfluenzae, Corynebacterium propinquum (#12), respectively (Table 1). None of the biopsies from the control group was found positive for biofilm-like structures (0/5). Figure 1G–H shows two representative examples of biopsies from control group (#1C and #2C).
CLSM imaging of sinus biopsies from patients #1, #4, #5, #6, #12 and controls #1C and #2C. The presence of bright green biofilm-like structures stained with Syto9 is evident in biopsies from CRS patients, but not from control patients. Aggregates of cocci shaped microorganisms (A–C–E). Aggregates of both cocci shaped microorganisms and long filamentous bacterial cells (B–D–F). Syto 9 staining (488/500–550 nm)
In few cases, multiple biopsy samples (2–5) were collected from CRS patients, in order to increase the sensitivity of the detection of the bacterial aggregates in the tissue, and individually evaluated by Syto9 staining and CLSM analysis. Of note, in one case in which it was possible to obtain 5 different biopsy samples, 3 of them were found positive for biofilm-like structures, while 2 of them were negative, suggesting that increasing the number of biopsies from a patient may increase the sensitivity of the technique.
A first analysis of the surface of the tissue sample from patient #4 resulted negative for the presence of bacterial aggregates. A successive more accurate analysis of the sample was performed by Z-stack imaging. Z-stack imaging revealed the presence of biofilm-like bacterial aggregates in the inner layers of sample #4 (Fig. 1B), indicating that biofilms may form not only on the mucosal surface, but also in the deeper layers of the tissue as also highlighted by others (Hall-Stoodley et al. 2012; Shields et al. 2013). For this reason, Z-stack imaging was performed also for all the other samples. Even with this accurate analysis no biofilm was detected in negative controls as well as in 4 of the CRS patients (#3, #7, #10, and #11) that were negative for the presence of biofilms on the surface of their biopsies (data not shown).
The bacterial aggregates detected deep in the tissue of patient #4 might also represent intramucosal microcolonies that have been also previously reported in CRS patients at a higher rate than in healthy controls (Wood et al. 2012; Kim et al. 2013a). Intramucosal bacteria were found to exhibit similar genotypic and phenotypic features as compared to surface bacteria isolated from the same patient and were hypothesized to reflect host mucosal immune dysfunction (Kim et al. 2013b).
Altogether, the results from CLSM analysis following fluorescent staining revealed the presence of bacterial aggregates resembling biofilms in the majority of the biopsies from CRS patients and in none of the control patients (P < 0.05). With regard to the staining protocol, Syto9 staining is considered a reliable method for biofilm detection, with excellent inter-observer reliability (Foreman et al. 2009). Furthermore, such method has the advantage of allowing the imaging of fresh tissues, thus not altering the biofilm architecture, and of requiring a short time (around 20 min) for sample preparation, making it suitable for analysis in routine microbiology laboratory.
Another method frequently used to detect biofilms on tissue samples is the CLSM associated with fluorescent in situ hybridization (FISH/CLSM), which employs specific probes to target complementary sequences of the microbial 16 s RNA (Mallmann et al. 2010; Lubbert et al. 2016). Depending on the probe used, this method allows the visualization of all the bacteria present (universal EUB probe) or the identification of single species of microorganisms (species-specific designed probe) forming the biofilm (Wecke et al. 2000; Mallmann et al. 2010). Although more accurate, FISH/CLSM technique is based on a time-consuming protocol requiring over 3 h to be completed (Foreman et al. 2009). Compared to this method, Syto9/CLSM is not species specific, but it gives a more rapid answer regarding biofilm presence or absence. For this reason, Syto9/CLSM has been proposed as the technique of choice for investigating the generic presence of biofilm in CRS patients, an information that could be of value for clinicians for its demonstrated impact on the post-operative course of CRS patients following endoscopic sinus surgery (Foreman et al. 2009; Singhal et al. 2010).
3.3 Isolation and Identification of Bacterial Strains from CRS Patients and Control Group by Microbiological Procedures
Diagnostic microbiological procedures aimed at the isolation of aerobic bacterial strains and fungi were adopted in this study. Overall, 34 different bacterial strains were isolated and identified (Table 1) from the clinical samples (11 nasal swabs, 11 sinus swabs, 4 aspirates and 17 tissue specimens) taken from the patients and the controls (Table S1). Clinical isolates belonged to a total of 11 different genera and 21 distinct species of Gram-positive (n°13 species) and Gram-negative (n° 8 species) bacteria.
Among the isolated microorganisms, the most frequently Gram-positive bacteria found in CRS patients belonged to the Staphylococcus genus with the following species distribution: S. aureus (3 strains), S. epidermidis (10 strains), S. lugdunensis (1 strain), S. haemolyticus (1 strain) and S. capitis (1 strain). These results are in agreement with previous culture-based investigations reporting a prevalence of staphylococci among strains isolated from clinical samples obtained from CRS patients (Brook 2006; Shields et al. 2013).
Almost one third of the isolated bacterial species were Gram-negative (e.g. Citrobacter koseri, Escherichia coli, Haemophilus parainfluenzae, H. paraphrophilus (recently renamed Aggregatibacter aphropilus), Moraxella catharralis, M. nonliquefaciens, Proteus mirabilis, Serratia liquefaciens). In two/third of the samples from CRS patients, more than one bacterial species was isolated (Table 1). S. liquefaciens, P. mirabilis, and E. coli, isolated from patients #2, #5 and #10 respectively, are rarely recovered from middle meatus samples from healthy individuals and therefore their recovery from symptomatic CRS patients suggests that they could play a pathologic role. As previously suggested (Brook 2011), these organisms may have been selected out following administration of antimicrobial therapy in patients with CRS. In particular, P. mirabilis was isolated from a patient who had suffered recurrent rhinosinusitis and numerous previous sinus surgeries. This observation is in accordance with a retrospective study by Brook and Franzier (2001), who found a correlation between the presence of Gram-negative bacteria in maxillary sinus aspirates and the history of previous sinus surgery in CRS patients. Serratia genus is not frequently found in community-acquired rhinosinusitis and, when isolated, S. marcescens is the most represented species isolated from nosocomial-acquired rhinosinusitis. In this study, S. liquefaciens was isolated from an immunocompetent patient (#2) who had no history of previous sinus surgery or long hospital stay. Extensive search in the literature revealed that S. liquefaciens is not a common Gram-negative bacteria found in CRS. Indeed, there are only three studies reporting the isolation of S. liquefaciens in rhinosinusitis (Snyman et al. 1988; Coffey et al. 2006; Richter and Gallagher 2016), and in one of them the bacterium was isolated from a case of acute sinusitis (Snyman et al. 1988).
Most of the bacteria isolated in this study were members of the oral or nasal normal flora. It is believed that the communication of the sinuses with the nasal cavity through the ostia could enable microorganisms that reside in the nasopharynx to spread into the sinus and, after closure of the ostia, these bacteria may be involved in the inflammation process (Brook 2011). No fungi were isolated from any of the specimens examined in this study.
The CRS study group included three subjects with nasal polyposis and nine patients without polyposis. Nasal polyps can impair paranasal sinus ventilation and drainage by blocking the ostiomeatal complex (Brook 2011). A recent study, based on the analysis of biopsies by fluorescent staining and CSLM, has shown that biofilms were more prevalent in CRS patients with nasal polyposis (33/34) compared to those without polyposis (22/27) and controls (14/25) (Danielsen et al. 2014). In our series, all the patients with polyposis (3/3) and 4/8 patients without polyposis presented biofilm-like aggregates in their biopsies. It has been reported that the microbial flora of the sinus of CRS patients with polyposis is not different from that found in patients with CRS without polyposis (Brook and Frazier 2001; Kim et al. 2014). In agreement with these studies, although biofilm seems to be more prevalent in CRS patients with polyposis, no evident correlation between the presence of polyposis and the severity of the disease was observed in our study group.
S. aureus, S. epidermidis and M. nonliquefaciens were isolated from nasal swabs and biopsies obtained from the control group subjects in agreement with the fact that such bacterial species may be part of the normal flora of nasal mucosa, although no evidence of biofilm-like structures was found in any of the subjects of our control group.
The question of whether normal bacterial flora in the sinuses exists is controversial (Brook 2011). Different studies have shown that the paranasal sinuses are sterile in healthy subjects with no history of sinus disease (Cook and Haber 1987; Sobin et al. 1992; Abou-Hamad et al. 2009). In contrast, other authors reported the presence of microorganisms in uninflamed sinus of healthy volunteers. In this regard, an early study in 1981 reported that S. pyogenes, S. aureus, S. pneumoniae, and H. influenzae can be commonly isolated from aspirates of patients without sign of CRS, who underwent corrective surgery for septal deviation (Brook 1981). Later, Jiang and colleagues endoscopically evaluated the bacterial flora of normal maxillary sinuses and isolated/cultured various microorganisms from half of the patients (Jiang et al. 1999). Recently, not only the presence of a normal flora, but also that of biofilms was demonstrated on the healthy mucosa of paranasal sinuses by scanning electron microscopy (Mladina et al. 2010). The discordance of the results among different studies might be due to: (i) the lack of standardization of the procedures used to collect samples from the sinus cavity; (ii) the failure to sterilize the area through which the endoscope is passed; (iii) the different anatomic areas from which the samples were collected (i.e. ethmoid bulla, maxillary antrum, or middle meatus); (iv) the kind of sample (swab, aspirate or mucosal tissue); (v) difference in the imaging or microbiological procedures adopted (e.g. transport time of the specimen, modalities of specimen processing and culturing). Further systematic studies with well-standardized protocols will be needed to clarify these aspects.
Regarding the procedures adopted in this study for the isolation of bacterial strains from surgical tissues, it is noteworthy that in 6/12 of the biopsies from CRS patients and in 1/5 of those from control subjects, the mere homogenization and vortexing of the tissue samples yielded negative culture results with no or very few colonies grown on agar plate (patients #1, #2, #4, #5, #6, #11, and #3C, Table 1). In contrast, isolation of bacterial strains was possible after incubating the same samples in an enrichment broth, suggesting that, as outlined by others, standard microbiological procedures might not be very efficient to detach bacteria embedded in the biofilm layers. Another hypothesis is that in vivo biofilm-forming bacteria are in a dormant/ametabolic/unculturable state and undergo re-activation only after inoculum in a rich liquid medium.
Different diagnostic procedures have been proposed to improve the isolation efficiency of bacteria from biofilms (Trampuz et al. 2007; Jost et al. 2014; Portillo et al. 2015). For instance, in the case a biofilm-associated joint prosthetic infections is suspected, sonication of orthopedic prostheses before the inoculum of the specimens into agar plates has demonstrated to promote the release of biofilm cells into planktonic form, increasing the number of positive cultures (Trampuz et al. 2007; Jost et al. 2014; Portillo et al. 2015). Very recently, Trampuz and coworkers have proved that the introduction of a sonication step and the inoculation of the sonication fluid into blood culture bottles further increases the isolation efficiency compared with the conventional sonication fluid or intraoperative tissue cultures (Portillo et al. 2015). It is likely that the introduction of a sonication step in the processing of biopsies taken from CRS patients for whom a biofilm infection is suspected could also improve the isolation efficiency directly from the tissue, avoiding the phase of the enrichment in broth. Indeed, although this latter step may facilitate the growth of bacteria embedded in the biofilm, in the case of mixed infections it may also alter the relative proportion of the bacterial species present in the sample, favoring those with faster growth kinetics (e.g. patient #10). On the other hand, the sonication step needs an accurate evaluation and standardization of the adopted parameters (e.g. duration and intensity of the treatment) to avoid loss of infectivity of the sample due to the sonication itself.
Routine antibiotic susceptibility testing was also performed on all the strains isolated from CRS patients and controls by semiautomatic systems. In most of the cases, the clinical isolates showed a wide range of antibiotic susceptibility (data not shown). The susceptibility profiles of S. aureus #5 and P. mirabilis #5 are reported in Table 3.
It is worth mentioning that a variety of molecular microbiological techniques has been developed in recent years that may help in the laboratory diagnosis of suspected biofilm infections with negative culture results (Costerton et al. 2011). Among them, particularly promising seems the combination of nucleic acid amplification procedures with electrospray ionization mass spectrometry (PCR/ESI-MS) (Ibis molecular method). Such technique, is based on the use of multiple set of primers designed to reveal known as well unknown bacteria. Detection and identification of fungi or viruses, or the presence in the specimen of bacterial genes that control the resistance to specific antibiotics (e.g. the MecA gene present in MRSA) is also possible. The amplicons produced by PCR are weighted by mass spectroscopy and their precise weight is used to calculate their base composition that is unique for each set of primers. Due to the high sensitivity and flexibility, the procedure has been proposed for the routine diagnosis of biofilm infections (Costerton et al. 2011).
3.4 Ability of Clinical Isolates to Form Biofilm In Vitro
A microtiter plate assay followed by CV staining (Brancatisano et al. 2014), was used to investigate in vitro the biofilm-forming ability of selected bacterial isolates that were classified into four categories, “no”, “weak”, “moderate”, and “strong” biofilm producer, as proposed by Stepanovic et al. (2007). The capacity of a microorganism to form a biofilm on a given surface is highly influenced by several factors including the nature of the surface or the environmental conditions (e.g. nutrients, pH, temperature) (Martin-Rodriguez et al. 2014). In order to evaluate the influence of the medium on biofilm-forming ability in vitro, we first compared the biofilm-forming ability of a selected group of clinical isolates in MHCA/Glu to that of bacteria grown in TSB/Glu medium. MHCA is usually used to test antibiotic susceptibility, but the majority of the standard biofilm formation assays are performed in TSB. No significant difference in biofilm formation was observed in the presence of TSB/Glu and MHCA/Glu (data not shown). Therefore, MHCA was chosen as a medium for the successive experiments.
Secondly, we compared the ability of selected strains to form biofilm in the presence of 1% glucose versus 1% sucrose. Sucrose is a fermentable disaccharide and, similarly to glucose, it serves as a substrate for the synthesis of the biofilm extracellular polysaccharide as it has been described especially for oral bacteria (Paes Leme et al. 2006). As shown in Table 2, for most, but not all of the tested strains, no evident difference was observed in biofilm-forming ability in the presence of 1% glucose versus 1% sucrose. Among the Gram-negative bacteria tested, S. liquefaciens #2 and P. mirabilis #5 were strong biofilm producers, while C. koseri #4 was a moderate producer in both media. Among the Gram-positive bacteria, S. anginosus #6 was a moderate producer in MHCA/Glu, while it was classified as a weak producer in MHCA/Suc. Both in MHCA/Glu and MHCA/Suc the three S. epidermidis strains isolated from CRS patients were moderate (#6, #7) or strong (#1) biofilm producers. The S. epidermidis ATCC35984 strain, used as a control, was confirmed as a strong producer. The S. epidermidis strain isolated from the control patient (#2C) was a moderate biofilm producer in both media. S. lugdunensis #4, another coagulase-negative Staphylococcus strain, also showed a moderate ability to produce biofilm in both media. Both S. aureus strains isolated from CRS patients (#3, #5) showed weak ability to produce biofilm in vitro, while the S. aureus ATCC 43300 strain, formed biofilm at a moderate level. The S. aureus strain isolated from the control patient (#1C) was a strong producer in MHCA/Glu, but a moderate producer in MHCA/Suc. Several studies have documented that ability of staphylococci to produce biofilm in vitro is not always fully expressed and often requires a modification of the growth broth, in order to be fully manifested (Arciola et al. 2002, 2015; Stepanovic et al. 2007).
For instance, supplementation of TSB with NaCl, which is a known activator of sigB gene locus regulating specific genes in stationary phase and under different stress conditions, led to increased biofilm formation and PIA synthesis by S. epidermidis 1457 (Knobloch et al. 2001). Moreover, in two different studies the rbf gene of S. aureus, a transcriptional regulator factor, was found to be involved in the positive regulation of the multicellular aggregation step of biofilm formation in response to glucose and NaCl (Lim et al. 2004; Cue et al. 2009). For these reasons, the ability of two strains of S. epidermidis (ATCC35984 and #1) and two strains of S. aureus (#3 and #5) to produce biofilm in presence of Gluc and/or NaCl was also tested. In the presence of 1% NaCl in the medium without glucose, a reduction of more than 50% in biofilm production was observed for the S. epidermidis ATCC35984 strain as compared to MHCA/Glu (data not shown), although the strain remained a strong biofilm producer (Table 2). A similar picture was obtained also for the S. epidermidis clinical isolate (#1), although the reduction was less evident (data not shown). In the presence of both glucose and NaCl, a reduction in biofilm production was visible for S. epidermidis strains ATCC35984. On the contrary in the same experimental conditions S. epidermidis #1 was still classified as strong biofilm producer. In the case of S. aureus (#3 and #5), adding of NaCl to the growth medium decreased the ability to form biofilm, both in the presence or absence of glucose.
It has been reported that S. aureus strains with poor ability to form biofilms in vitro, may express marked biofilm-forming capacity in vivo (Cardile et al. 2014). Factors potentially causing discrepancies between in vivo and in vitro conditions include the presence of host proteins, such as those present in human plasma (Aly and Levit 1987; Vaudaux et al. 1989; Wagner et al. 2011; Bjarnsholt et al. 2013). Body fluids may contain plasma with variable protein content according to the body district. For example, in nasal secretions, the plasma protein concentration ranges from 15 to 45% (Cardile et al. 2014) The importance of host proteins in facilitating biofilm formation is highlighted by studies demonstrating that medical implants are often coated by various host matrix proteins that enhance bacterial attachment to the surface and biofilm formation in vivo (Wagner et al. 2011; Bjarnsholt et al. 2013). In vitro the presence of plasma has been shown to promote biofilm growth as well (Chen et al. 2012; Walker and Horswill 2012). For this reason, the effect of 1% human plasma on biofilm formation was tested (Fig. 2, Table 2). Most of the tested S. aureus strains increased the production of biofilm in the presence of human plasma, passing from weak to moderate biofilm producers or, in the case of the S. aureus ATCC strain, from moderate to strong producer according to the categories of Stepanovic. Only the S. aureus #1C, isolated from one of the control patients, showed a slight reduction in biofilm formation but remained a strong biofilm producer (Fig. 2). A similar biofilm-inducing effect was not seen for the coagulase-negative staphylococcal strains (S. epidermidis ATCC35984 and #1, and S. lugdunensis #4) and for all the other strains tested (Fig. 2, Table 2). These data support the hypothesis that the supplementation of media with human plasma may better mimic the conditions encountered in vivo by certain, but not all the strains, favoring their ability to form biofilms in vitro.
In vitro biofilm production by clinical isolates and ATCC strains of S. epidermidis and S. aureus in MHCA/Glucose or in MHCA/Glucose added with 1% of inactivated human plasma. For each strain the biofilm biomass was evaluated after 24 h of incubation at 37 °C by CV staining and measurement of the optical density (OD) at 595 nm. Horizontal lines indicate the OD cut-off values used to define the categories of biofilm producers according to Stepanovic et al. (2007). OD values higher than the continuous line indicate strong biofilm producers
The ability of a clinical isolate to form biofilm in vitro has been proposed as a predictive marker of its capacity to form biofilm also in vivo (Sanchez et al. 2013). Rapid and simple procedures for the routine analysis of biofilm forming ability of clinical isolates in the clinical microbiology laboratory have been recently proposed, with the aim to help the clinicians to identify high-risk infections and/or to predict the risk of therapeutic failure (Di Domenico et al. 2016). The results presented herein suggest caution in this respect, as they demonstrate that the ability to form biofilm in vitro might be highly influenced by the growing conditions adopted. Thus, poor ability to form biofilm in vitro, not necessarily is an indication of a low propensity of a strain to form biofilm also in vivo, but rather could indicate that the conditions adopted in vitro to grow that strain are sub-optimal.
3.5 Study of 3D Architecture and Extracellular Matrix Composition of Biofilms Formed In Vitro by Clinical Isolates
A more detailed characterization of biofilms formed in vitro by selected bacterial strains isolated from CRS patients was carried out after 24 h of growth in MHCA added with 1% glucose or, in the case of S. aureus, with 1% glucose and 1% human plasma. Figure 3A shows some representative CLSM images of biofilms stained with Syto9. The typical 3D architecture of biofilms, in which groups of microbial cells are separated by open water channels for the delivery of nutrients and oxygen, and the removal of metabolic waste is evident for most of the strains analyzed. In few cases, a peculiar pattern of bacterial distribution or cell morphology was observed as in case of S. liquefaciens. In this case, characteristic filamentous cells, longer than their planktonic counterparts, were visible in agreement to what previously reported for the MG1 strain of S. liquefaciens (Givskov et al. 1998).
In vitro phenotypic characterisation of biofilms produced by different clinical strains. (A) Architecture of biofilms produced in vitro by clinical isolates from CRS patients as visualized by Syto9 staining (488/500–550 nm) and CLSM analysis. Strains were cultivated in MHCA added with 1% glucose and, in the case of S. aureus, also with 1% human plasma. (B) Visualisation of biofilm matrix components of S. aureus and S. epidermidis clinical isolates by CLSM and double staining with WGA488 (detection of PIA in the green channel) and SpyroRuby (detection of proteins in the red channel). Scale bar: 10 μm
One of the main components of biofilms is the EPS, an abundant and complex matrix constituted by exopolysaccharides, proteins, extracellular DNA and other components that surround and protect biofilm cells (Donlan and Costerton 2002). The most characterized exopolysaccharide matrix component of S. aureus and S. epidermidis biofilms is PIA, also known as poly-N-acetyl-glucosamine (Arciola et al. 2015). When produced, PIA mediates adhesion to surfaces acting as a cementing matrix, enabling bacterial cells to agglomerate in multi-layered biofilms, and making bacteria less accessible to the host defence system and antibiotics (Arciola et al. 2002). In order to phenotypically investigate the production of PIA by Staphylococcus strains isolated in our study, the colony morphology of such strains was analysed by the Congo Red Agar (CRA) plate method (Freeman et al. 1989). When grown on CRA, all the staphylococcal strains tested, but one, developed “black/gray” colonies indicative of a PIA-producing ability (see Fig. 4 for examples of colony morphology). The only staphylococcal strain that formed red colonies on CRA was S. lugdunensis, in agreement with the reported inability of this species to produce PIA (Arciola et al. 2015; Ravaioli et al. 2012). It has been reported that expression of PIA in S. epidermidis is subjected to phase variation due to the reversible insertion of an IS element in the operon encoding for PIA (Ziebuhr et al. 1999), although this mechanism has not been confirmed in later studies (Arciola et al. 2004). Phase variation of PIA expression has been described also for S. aureus. In S. aureus the phenomenon seems to be due to an expansion of the tetranucleotide tandem repeat housed in the PIA operon, resulting in a PIA-negative biofilm (Brooks and Jefferson 2014). It has been proposed that the ability to rapidly switch between phenotypes allows staphylococci to adapt to changing environmental conditions conferring an evolutionary advantage in the relationship with their hosts (Arciola et al. 2015).
Colony morphology on Congo red agar plates after 24 h incubation at 37 °C followed by 24 h incubation at room temperature. PIA producing ability was evaluated by visual detection of the colony color according to the chromatic scale of Arciola et al. (2002). Two PIA-positive strains (S. epidermidis #1 and S. aureus #3) and one PIA-negative strain (S. lugdunensis #4) are shown
In order to further characterise the biofilms of the staphylococcal strains isolated in this study in terms of PIA and/protein production, CLSM imaging of S. aureus and S. epidermidis biofilms was performed following double staining with the wheat germ agglutinin-Oregon green 488 conjugate (WGA488) or with the red fluorescent dye Sypro Ruby to label PIA and proteins, respectively (Fig. 3B). In agreement with the results obtained by using the CRA method, all biofilms appeared green due to WGA488 binding to the N-acetyl-glucosamine residues of PIA in their matrix. Moreover, red fluorescence, due to Sypro Ruby binding, was also detectable in the biofilms of some strains (e.g. S. epidermidis #6, S. aureus #3, Fig. 3B), indicating the presence of protein components in the biofilm matrix of these strains. Interestingly, by using proteomic approaches, recently Gil and coworkers characterized the exoproteome of exopolysaccharide-based and protein-based biofilm matrices produced by two clinical isolates of S. aureus (Gil et al. 2014). They found that independently of the nature of the biofilm matrix, a common set of secreted proteins is contained in both types of exoproteomes.
A number of proteins localized in the extracellular matrix of biofilms have been described to be involved in generating PIA-independent biofilms. These include the biofilm-associated protein (Bap), a 2276-amino acid surface protein, and the accumulated associated protein (Aap) (Arciola et al. 2015). In addition, some authors suggest that various surface proteins called cell-wall anchored (CWA) proteins (including SasG, SasC, Protein A, FnBPA) can contribute to mediate the initial attachment and biofilm accumulation and maturation in Staphylococcus spp. (Merino et al. 2009; Vergara-Irigaray et al. 2009; Speziale et al. 2014). It has been proposed that staphylococci may produce PIA-dependent or PIA-independent biofilms in different phases of the pathogenesis of an infection and that this ability may allow the bacteria to better adapt to the multiple stimuli encountered in the host.
3.6 Antibiotic Susceptibility Testing of Bacterial Isolates in Biofilm Form
Several studies have indicated that activity of most antibiotics shows significant quantitative and qualitative differences against biofilm bacteria as compared to their planktonic counterparts. Therefore, it has been proposed that, when a biofilm-associated infection is suspected, the evaluation of antimicrobial susceptibility in biofilm form may better predict the success of the therapy and offers clinicians more appropriate guidelines to treat such infection than standard antimicrobial susceptibility assays (Macia et al. 2014; Hoiby et al. 2015). Although various methods have been described for testing and quantifying the activity of antibiotics against sessile bacteria over the last decade (Macia et al. 2014), to date, none of them have been approved as reference method in the clinical microbiology laboratory and there is a growing interest in the development of new susceptibility tests specific for biofilm-growing bacteria.
In a first set of experiments of this study, the colorimetric AB cell viability assay (Pettit et al. 2005) was used to test the antibiotic susceptibility of pre-formed biofilms of two Gram-positive (S. aureus #5 and S. epidermidis #1), and two Gram-negative (S. liquefaciens #2 and P. mirabilis #5) bacteria isolated from CRS-patients. Such assay is based on the evaluation of bacterial viability within the biofilm by the use of the blue dye resazurin. Reduction of the redox indicator by viable bacteria causes a colour change from non-fluorescent, blue (oxidized form) to fluorescent, red (reduced form) (Pettit et al. 2005; Tote et al. 2008). Two-fold serial dilutions of different antibiotics acting with distinct mode of actions were added to pre-formed (24 h–old) biofilms and incubated at 37 °C for 24 h. Then, AB was added to the wells and, after 1 h of incubation at 37 °C the MBIC was determined visually. MBIC was defined as the lowest drug concentration resulting in at least 50% reduction of AB that corresponded visually to a purplish coloured well 60 min after the addition of AB (Pettit et al. 2005).
As shown in Table 3 for S. aureus #5 and P. mirabilis #5, overall MBIC values against biofilms of all tested strains were considerably higher than those obtained against planktonic bacteria by routine semi-automated antibiotic susceptibility testing. Of note, most of the antibiotics tested were not able to inhibit the metabolic activity of biofilms even at concentrations as high as 1024 μg/ml (Table 3).
Biofilm recalcitrance to antimicrobial treatment is a multifactorial phenomenon (Lebeaux et al. 2014) where inability of certain antibiotics to diffuse through the biofilm extracellular matrix may play a significant role. According to the charge and other physical-chemical features of both the antibiotic and the extracellular matrix, the antibiotic might be repulsed or alternatively sequestered by matrix components hampering its penetration into the deeper biofilm layers (Lewis 2008; Lebeaux et al. 2014; Macia et al. 2014). Rifampicin was the most active antibiotic among those tested against staphylococcal biofilms, in agreement with the previously reported ability of such antibiotic to penetrate through the biofilm extracellular matrix (Dunne et al. 1993; Zheng and Stewart 2002). However, due to the rapid development of the resistance against rifampicin by staphylococcal strains, its use against biofilms is not recommended as mono-therapy, but in combination with other antimicrobials including tigecycline (Trampuz and Zimmerli 2006; Aslam and Darouiche 2007), linezolid (Raad et al. 1998; Saginur et al. 2006) and ciprofloxacin (Widmer et al. 1992; Zimmerli et al. 1998). The efficacy of the rifampicin against staphylococcal biofilms may suggest that targeting RNA polymerase is a highly effective strategy against the metabolic diversity of cells found in biofilms (Fey 2010).
Overall, a correlation was observed between the ability of an antibiotic to diffuse through the extracellular matrix and its ability to treat mature biofilms. For instance, although the strain of S. aureus tested (#5) was sensitive to vancomycin in planktonic form, biofilms of the same strain were highly resistant to vancomycin (Table 3) in agreement with the low capacity of this antibiotic to penetrate S. aureus biofilms (Jefferson et al. 2005). Fluoroquinolones are reported to penetrate well into the extracellular matrix of biofilms formed by Gram negative species such as P. aeruginosa and E. coli (Lebeaux et al. 2014). In accordance, levofloxacin exhibited an antibiofilm activity at low concentrations (MBIC ≤ 4 μg/ml) against P. mirabilis tested in this study (Table 3). Doxycycline was able to inhibit biofilm metabolic activity of the same strain at a relatively low concentration (MBIC = 128 μg/ml), even if higher than that achievable in clinical practice.
IMC is a highly sensitive technique, which allows measuring the heat flow generated or consumed during a biological process such as bacterial growth (Braissant et al. 2010). IMC has been previously used with success for clinical and diagnostic purposes to test the activity of antibiotics and antimycotics against bacterial and fungal biofilms (Furustrand Tafin et al. 2012; Furustrand Tafin et al. 2013; Oliva et al. 2014). In this study, IMC was used to evaluate the MBEC of some of the antibiotics previously tested against S. aureus #5 and P. mirabilis #5 by AB assay, (Fig. 5 and Table 3).
Determination of the minimal biofilm eradicating concentration (MBEC) by isothermal microcalorimetry. 24 h-old biofilms, grown on porous glass beads, were exposed to different concentrations of various antibiotics. Beads were then washed to remove non-attached bacteria as well as antibiotics and incubated in the microcalorimeter in fresh medium (without antibiotics). The curves represent the heat flow produced over time by S. aureus #5 (A–F) and P. mirabilis #5 (G–J). The numbers above each curve indicate the antibiotic concentrations tested expressed in μg/ml. The circled value is the minimum biofilm eradication concentration (MBEC), and corresponds to a flat line indicative of a lack of regrowth. PC: positive control, bacterial biofilms without any pre-treatment with antibiotics. NC: negative control, medium containing sterile beads
Twenty-four hour-old biofilms grown on porous glass beads were incubated in the presence of two-fold serial dilutions of each antibiotic. Ability of biofilm cells to re-grow after antibiotic treatment was evaluated by IMC incubating beads in antibiotic-free medium. Figure 5 depicts the curves obtained at each antibiotic concentration tested, plotted as heat flow (in microwatt) versus time (h). With the exception of vancomycin and erythromycin, which did not show complete inhibition of heat production even at concentrations up to 1024 μg/ml, a complete suppression of heat production was observed in presence of 128 μg/ml rifampicin, 256 μg/ml daptomycin, 64 μg/ml doxycycline, and 1024 μg/ml levofloxacin against S. aureus #5 (Fig. 5, Table 3). Amikacin inhibited the heat production by P. mirabilis #5 at 256 μg/ml, and levofloxacin and doxycycline at 512 μg/ml. On the contrary, amoxicillin up to 1024 μg/ml, did not abolish heat production by the sessile P. mirabilis (Fig. 5, Table 3).
4 Conclusions
Aggregates of bacteria resembling biofilms were detected in the majority of the biopsies of CRS patients. Syto9 staining of the tissue specimens followed by CLSM analysis revealed to be an easy to perform and rapid technique with the potential to be employed in adequately equipped routine microbiology laboratory. The use of more detailed analyses, such as the Z-stack technique, helped to visualise biofilms not only on the tissue surface, but also in the deeper layers of the biopsy, increasing the detection sensitivity. The sampling of multiple biopsies from various locations of the inflamed mucosa could further increase the probability to detect the presence of biofilms.
Interestingly, biofilm-like structures were not detected in any of the control patients, although bacterial strains potentially part of the normal flora were also isolated from biopsies of such subjects. This may suggest that the mere presence of bacteria in the sinus mucosa does not necessarily leads to pathology. It might be possible that in individuals with specific anatomical features or other predisposing factors bacteria originating from the upper respiratory tract or oral cavity may stably colonize the sinus mucosa and form biofilms, thus contributing to the chronicisation of the infection.
The consistent demonstration of biofilms on mucosa biopsies in CRS patients suggests that they may play a role in the pathogenesis/persistence of the disease.
Isolation of biofilm-forming bacteria from biopsies may be troublesome and require appropriate and standardized procedures such as inoculation in an enrichment broth or sonication to allow bacteria to detach from the tissue. Most of the bacterial species isolated from biopsies of CRS patients, belonged to the normal flora of the upper-respiratory tract or oral cavity, while in few cases the isolated bacteria belonged to the Enterobacteriaceae.
The type of medium used in the biofilm-forming assay in vitro had an impact on the ability to form biofilm of bacterial strains isolated from biopsies of CRS patients and controls. Overall, rich media, resembling the in vivo milieu, and containing high glucose concentrations, human plasma, and/or salts, promoted biofilm formation, but not for all the strains. When tested in their optimized growth conditions the large majority of the strains isolated from biopsy samples were moderate or strong biofilm producers in vitro.
When a biofilm-associated infection is suspected, antibiotic susceptibility testing against biofilms of the clinical strains isolated from the infection site could be more appropriate than the standard assays performed on planktonic bacteria, to guide the therapy. Nevertheless, standardization of the procedures, parameters and breakpoints, by official agencies, is needed before they are implemented in the clinical microbiology laboratories for routine susceptibility testing.
The introduction of innovative techniques such as isothermal microcalorimetry in the microbiology laboratory may help to perform large scale and accurate antibiotic susceptibility testing against bacteria in biofilms.
References
Abou-Hamad W, Matar N, Elias M, Nasr M, Sarkis-Karam D, Hokayem N, Haddad A (2009) Bacterial flora in normal adult maxillary sinuses. Am J Rhinol Allergy 23(3):261–263. doi:10.2500/ajra.2009.23.3317
Aly R, Levit S (1987) Adherence of Staphylococcus aureus to squamous epithelium: role of fibronectin and teichoic acid. Rev Infect Dis 9(Suppl 4):S341–S350
Arciola CR, Campoccia D, Gamberini S, Cervellati M, Donati E, Montanaro L (2002) Detection of slime production by means of an optimised Congo red agar plate test based on a colourimetric scale in Staphylococcus epidermidis clinical isolates genotyped for ica locus. Biomaterials 23 (21):4233–4239. doi:Pii S0142–9612(02)00171–0. doi:10.1016/S0142-9612(02)00171-0
Arciola CR, Campoccia D, Gamberini S, Rizzi S, Donati ME, Baldassarri L, Montanaro L (2004) Search for the insertion element IS256 within the ica locus of Staphylococcus epidermidis clinical isolates collected from biomaterial-associated infections. Biomaterials 25(18):4117–4125. doi:10.1016/j.biomaterials.2003.11.027
Arciola CR, Campoccia D, Ravaioli S, Montanaro L (2015) Polysaccharide intercellular adhesin in biofilm: structural and regulatory aspects. Front Cell Infect Mi 5. doi:ARTN 7. 10.3389/fcimb.2015.00007
Aslam S, Darouiche RO (2007) Prolonged bacterial exposure to minocycline/rifampicin-impregnated vascular catheters does not affect antimicrobial activity of catheters. J Antimicrob Chemother 60(1):148–151. doi:10.1093/jac/dkm173
Batoni G, Maisetta G, Esin S (2016) Antimicrobial peptides and their interaction with biofilms of medically relevant bacteria. Biochim Biophys Acta 1858(5):1044–1060. doi:10.1016/j.bbamem.2015.10.013
Benninger MS, Ferguson BJ, Hadley JA, Hamilos DL, Jacobs M, Kennedy DW, Lanza DC, Marple BF, Osguthorpe JD, Stankiewicz JA, Anon J, Denneny J, Emanuel I, Levine H (2003) Adult chronic rhinosinusitis: definitions, diagnosis, epidemiology, and pathophysiology. Otolaryngol Head Neck Surg 129(3 Suppl):S1–32
Bjarnsholt T, Alhede M, Alhede M, Eickhardt-Sorensen SR, Moser C, Kuhl M, Jensen PO, Hoiby N (2013) The in vivo biofilm. Trends Microbiol 21(9):466–474. doi:10.1016/j.tim.2013.06.002
Braissant O, Wirz D, Gopfert B, Daniels AU (2010) Use of isothermal microcalorimetry to monitor microbial activities. FEMS Microbiol Lett 303(1):1–8. doi:10.1111/j.1574-6968.2009.01819.x
Brancatisano FL, Maisetta G, Di Luca M, Esin S, Bottai D, Bizzarri R, Campa M, Batoni G (2014) Inhibitory effect of the human liver-derived antimicrobial peptide hepcidin 20 on biofilms of polysaccharide intercellular adhesin (PIA)-positive and PIA-negative strains of Staphylococcus epidermidis. Biofouling 30(4):435–446. doi:10.1080/08927014.2014.888062
Brook I (1981) Bacteriologic features of chronic sinusitis in children. JAMA 246(9):967–969
Brook I (2006) Bacteriology of chronic sinusitis and acute exacerbation of chronic sinusitis. Arch Otolaryngol Head Neck Surg 132(10):1099–1101. doi:10.1001/archotol.132.10.1099
Brook I (2011) Microbiology of sinusitis. Proc Am Thorac Soc 8(1):90–100. doi:10.1513/pats.201006-038RN
Brook I, Frazier EH (2001) Correlation between microbiology and previous sinus surgery in patients with chronic maxillary sinusitis. Ann Otol Rhinol Laryngol 110(2):148–151
Brooks JL, Jefferson KK (2014) Phase variation of poly-N-acetylglucosamine expression in Staphylococcus aureus. PLoS Pathog 10(7):e1004292. doi:10.1371/journal.ppat.1004292
Cardile AP, Sanchez CJ Jr, Samberg ME, Romano DR, Hardy SK, Wenke JC, Murray CK, Akers KS (2014) Human plasma enhances the expression of Staphylococcal microbial surface components recognizing adhesive matrix molecules promoting biofilm formation and increases antimicrobial tolerance in vitro. BMC Res Notes 7:457. doi:10.1186/1756-0500-7-457
Chen P, Abercrombie JJ, Jeffrey NR, Leung KP (2012) An improved medium for growing Staphylococcus aureus biofilm. J Microbiol Methods 90(2):115–118. doi:10.1016/j.mimet.2012.04.009
Coffey CS, Sonnenburg RE, Melroy CT, Dubin MG, Senior BA (2006) Endoscopically guided aerobic cultures in postsurgical patients with chronic rhinosinusitis. Am J Rhinol 20(1):72–76
Cook HE, Haber J (1987) Bacteriology of the maxillary sinus. J Oral Maxillofac Surg 45(12):1011–1014
Costerton JW, Post JC, Ehrlich GD, Hu FZ, Kreft R, Nistico L, Kathju S, Stoodley P, Hall-Stoodley L, Maale G, James G, Sotereanos N, DeMeo P (2011) New methods for the detection of orthopedic and other biofilm infections. FEMS Immunol Med Microbiol 61(2):133–140. doi:10.1111/j.1574-695X.2010.00766.x
Cue D, Lei MG, Luong TT, Kuechenmeister L, Dunman PM, O'Donnell S, Rowe S, O'Gara JP, Lee CY (2009) Rbf promotes biofilm formation by Staphylococcus Aureus via repression of icaR, a negative regulator of icaADBC. J Bacteriol 191(20):6363–6373. doi:10.1128/jb.00913-09
Danielsen KA, Eskeland O, Fridrich-Aas K, Orszagh VC, Bachmann-Harildstad G, Burum-Auensen E (2014) Bacterial biofilms in patients with chronic rhinosinusitis: a confocal scanning laser microscopy study. Rhinology 52(2):150–155. doi:10.4193/Rhin
Desrosiers M, Evans GA, Keith PK, Wright ED, Kaplan A, Bouchard J, Ciavarella A, Doyle PW, Javer AR, Leith ES, Mukherji A, Robert Schellenberg R, Small P, Witterick IJ (2011) Canadian clinical practice guidelines for acute and chronic rhinosinusitis. J Otolaryngol Head Neck Surg 40(Suppl 2):S99–193
Di Domenico EG, Toma L, Provot C, Ascenzioni F, Sperduti I, Prignano G, Gallo MT, Pimpinelli F, Bordignon V, Bernardi T, Ensoli F (2016) Development of an in vitro assay, based on the BioFilm ring test(R), for rapid profiling of biofilm-growing bacteria. Front Microbiol 7:1429. doi:10.3389/fmicb.2016.01429
Donlan RM, Costerton JW (2002) Biofilms: survival mechanisms of clinically relevant microorganisms. Clin Microbiol Rev 15(2):167–193
Dunne WM Jr, Mason EO Jr, Kaplan SL (1993) Diffusion of rifampin and vancomycin through a Staphylococcus epidermidis biofilm. Antimicrob Agents Chemother 37(12):2522–2526
Fey PD (2010) Modality of bacterial growth presents unique targets: how do we treat biofilm-mediated infections? Curr Opin Microbiol 13(5):610–615. doi:10.1016/j.mib.2010.09.007
Fokkens WJ, Lund VJ, Mullol J, Bachert C, Alobid I, Baroody F, Cohen N, Cervin A, Douglas R, Gevaert P, Georgalas C, Goossens H, Harvey R, Hellings P, Hopkins C, Jones N, Joos G, Kalogjera L, Kern B, Kowalski M, Price D, Riechelmann H, Schlosser R, Senior B, Thomas M, Toskala E, Voegels R, de Wang Y, Wormald PJ (2012) European Position Paper on Rhinosinusitis and Nasal Polyps 2012. Rhinol Suppl (23):3 p preceding table of contents, 1–298
Foreman A, Psaltis AJ, Tan LW, Wormald PJ (2009) Characterization of bacterial and fungal biofilms in chronic rhinosinusitis. Am J Rhinol Allergy 23(6):556–561. doi:10.2500/ajra.2009.23.3413
Foreman A, Singhal D, Psaltis AJ, Wormald PJ (2010) Targeted imaging modality selection for bacterial biofilms in chronic rhinosinusitis. Laryngoscope 120(2):427–431. doi:10.1002/lary.20705
Freeman DJ, Falkiner FR, Keane CT (1989) New method for detecting slime production by coagulase negative staphylococci. J Clin Pathol 42(8):872–874
Furustrand Tafin U, Meis JF, Trampuz A (2012) Isothermal microcalorimetry for antifungal susceptibility testing of Mucorales, Fusarium spp., and Scedosporium spp. Diagn Microbiol Infect Dis 73(4):330–337. doi:10.1016/j.diagmicrobio.2012.05.009
Furustrand Tafin U, Orasch C, Trampuz A (2013) Activity of antifungal combinations against Aspergillus species evaluated by isothermal microcalorimetry. Diagn Microbiol Infect Dis 77(1):31–36. doi:10.1016/j.diagmicrobio.2013.06.004
Gil C, Solano C, Burgui S, Latasa C, Garcia B, Toledo-Arana A, Lasa I, Valle J (2014) Biofilm matrix exoproteins induce a protective immune response against Staphylococcus aureus biofilm infection. Infect Immun 82(3):1017–1029. doi:10.1128/iai.01419-13
Givskov M, Ostling J, Eberl L, Lindum PW, Christensen AB, Christiansen G, Molin S, Kjelleberg S (1998) Two separate regulatory systems participate in control of swarming motility of Serratia liquefaciens MG1. J Bacteriol 180(3):742–745
Hall-Stoodley L, Stoodley P, Kathju S, Hoiby N, Moser C, Costerton JW, Moter A, Bjarnsholt T (2012) Towards diagnostic guidelines for biofilm-associated infections. FEMS Immunol Med Microbiol 65(2):127–145. doi:10.1111/j.1574-695X.2012.00968.x
Hoiby N, Bjarnsholt T, Moser C, Bassi GL, Coenye T, Donelli G, Hall-Stoodley L, Hola V, Imbert C, Kirketerp-Moller K, Lebeaux D, Oliver A, Ullmann AJ, Williams C, Biofilms ESGf, Consulting External Expert, Werner Z (2015) ESCMID guideline for the diagnosis and treatment of biofilm infections 2014. Clin Microbiol Infect 21(Suppl 1):S1–25. doi:10.1016/j.cmi.2014.10.024
Jefferson KK, Goldmann DA, Pier GB (2005) Use of confocal microscopy to analyze the rate of vancomycin penetration through Staphylococcus aureus biofilms. Antimicrob Agents Chemother 49(6):2467–2473. doi:10.1128/aac.49.6.2467-2473.2005
Jiang RS, Liang KL, Jang JW, Hsu CY (1999) Bacteriology of endoscopically normal maxillary sinuses. J Laryngol Otol 113(9):825–828
Jost GF, Wasner M, Taub E, Walti L, Mariani L, Trampuz A (2014) Sonication of catheter tips for improved detection of microorganisms on external ventricular drains and ventriculo-peritoneal shunts. J Clin Neurosci 21(4):578–582. doi:10.1016/j.jocn.2013.05.025
Kim R, Freeman J, Waldvogel-Thurlow S, Roberts S, Douglas R (2013a) The characteristics of intramucosal bacteria in chronic rhinosinusitis: a prospective cross-sectional analysis. Int Forum Allergy Rhinol 3(5):349–354. doi:10.1002/alr.21117
Kim RJ, Yin T, Chen CJ, Mansell CJ, Wood A, Dunbar PR, Douglas RG (2013b) The interaction between bacteria and mucosal immunity in chronic rhinosinusitis: a prospective cross-sectional analysis. Am J Rhinol Allergy 27(6):e183–e189. doi:10.2500/ajra.2013.27.3974
Kim YM, Jin J, Choi JA, Cho SN, Lim YJ, Lee JH, Seo JY, Chen HY, Rha KS, Song CH (2014) Staphylococcus aureus enterotoxin B-induced endoplasmic reticulum stress response is associated with chronic rhinosinusitis with nasal polyposis. Clin Biochem 47(1–2):96–103. doi:10.1016/j.clinbiochem.2013.10.030
Knobloch JK, Bartscht K, Sabottke A, Rohde H, Feucht HH, Mack D (2001) Biofilm formation by Staphylococcus epidermidis depends on functional RsbU, an activator of the sigB operon: differential activation mechanisms due to ethanol and salt stress. J Bacteriol 183(8):2624–2633. doi:10.1128/JB.183.8.2624-2633.2001
Lam K, Schleimer R, Kern RC (2015) The etiology and pathogenesis of chronic Rhinosinusitis: a review of current hypotheses. Curr Allergy Asthma Rep 15(7):41. doi:10.1007/s11882-015-0540-2
Lanza DC, Kennedy DW (1997) Adult rhinosinusitis defined. Otolaryngol Head Neck Surg 117(3 Pt 2):S1–S7
Lebeaux D, Ghigo JM, Beloin C (2014) Biofilm-related infections: bridging the gap between clinical management and fundamental aspects of recalcitrance toward antibiotics. Microbiol Mol Biol Rev 78(3):510–543. doi:10.1128/MMBR.00013-14
Lewis K (2008) Multidrug tolerance of biofilms and persister cells. Curr Top Microbiol Immunol 322:107–131
Lim Y, Jana M, Luong TT, Lee CY (2004) Control of glucose- and NaCl-induced biofilm formation by rbf in Staphylococcus aureus. J Bacteriol 186(3):722–729
Lubbert C, Wendt K, Feisthammel J, Moter A, Lippmann N, Busch T, Mossner J, Hoffmeister A, Rodloff AC (2016) Epidemiology and resistance patterns of bacterial and fungal colonization of biliary plastic stents: a prospective cohort study. PLoS One 11(5):e0155479. doi:10.1371/journal.pone.0155479
Macia MD, Rojo-Molinero E, Oliver A (2014) Antimicrobial susceptibility testing in biofilm-growing bacteria. Clin Microbiol Infect 20(10):981–990. doi:10.1111/1469-0691.12651
Maisetta G, Grassi L, Di Luca M, Bombardelli S, Medici C, Brancatisano FL, Esin S, Batoni G (2016) Anti-biofilm properties of the antimicrobial peptide temporin 1Tb and its ability, in combination with EDTA, to eradicate Staphylococcus epidermidis biofilms on silicone catheters. Biofouling 32(7):787–800. doi:10.1080/08927014.2016.1194401
Mallmann C, Siemoneit S, Schmiedel D, Petrich A, Gescher DM, Halle E, Musci M, Hetzer R, Gobel UB, Moter A (2010) Fluorescence in situ hybridization to improve the diagnosis of endocarditis: a pilot study. Clin Microbiol Infect 16(6):767–773. doi:10.1111/j.1469-0691.2009.02936.x
Martin-Rodriguez AJ, Gonzalez-Orive A, Hernandez-Creus A, Morales A, Dorta-Guerra R, Norte M, Martin VS, Fernandez JJ (2014) On the influence of the culture conditions in bacterial antifouling bioassays and biofilm properties: Shewanella algae, a case study. BMC Microbiol 14:102. doi:10.1186/1471-2180-14-102
Merino N, Toledo-Arana A, Vergara-Irigaray M, Valle J, Solano C, Calvo E, Lopez JA, Foster TJ, Penades JR, Lasa I (2009) Protein A-mediated multicellular behavior in Staphylococcus Aureus. J Bacteriol 191(3):832–843. doi:10.1128/jb.01222-08
Mladina R, Skitarelic N, Music S, Ristic M (2010) A biofilm exists on healthy mucosa of the paranasal sinuses: a prospectively performed, blinded, scanning electron microscope study. Clin Otolaryngol 35(2):104–110. doi:10.1111/j.1749-4486.2010.02097.x
Oliva A, Furustrand Tafin U, Maiolo EM, Jeddari S, Betrisey B, Trampuz A (2014) Activities of fosfomycin and rifampin on planktonic and adherent Enterococcus faecalis strains in an experimental foreign-body infection model. Antimicrob Agents Chemother 58(3):1284–1293. doi:10.1128/AAC.02583-12
Paes Leme AF, Koo H, Bellato CM, Bedi G, Cury JA (2006) The role of sucrose in cariogenic dental biofilm formation--new insight. J Dent Res 85(10):878–887
Penesyan A, Gillings M, Paulsen IT (2015) Antibiotic discovery: combatting bacterial resistance in cells and in biofilm communities. Molecules 20(4):5286–5298. doi:10.3390/molecules20045286
Peters AT, Spector S, Hsu J, Hamilos DL, Baroody FM, Chandra RK, Grammer LC, Kennedy DW, Cohen NA, Kaliner MA, Wald ER, Karagianis A, Slavin RG, Joint Task Force on Practice Parameters rtAAoAA, Immunology tACoAA, Immunology, the Joint Council of Allergy A, Immunology (2014) Diagnosis and management of rhinosinusitis: a practice parameter update. Ann Allergy Asthma Immunol 113(4):347–385. doi:10.1016/j.anai.2014.07.025
Pettit RK, Weber CA, Kean MJ, Hoffmann H, Pettit GR, Tan R, Franks KS, Horton ML (2005) Microplate Alamar blue assay for Staphylococcus epidermidis biofilm susceptibility testing. Antimicrob Agents Chemother 49(7):2612–2617. doi:10.1128/AAC.49.7.2612-2617.2005
Portillo ME, Salvado M, Trampuz A, Siverio A, Alier A, Sorli L, Martinez S, Perez-Prieto D, Horcajada JP, Puig-Verdie L (2015) Improved diagnosis of orthopedic implant-associated infection by inoculation of sonication fluid into blood culture bottles. J Clin Microbiol 53(5):1622–1627. doi:10.1128/JCM.03683-14
Psaltis AJ, Ha KR, Beule AG, Tan LW, Wormald PJ (2007) Confocal scanning laser microscopy evidence of biofilms in patients with chronic rhinosinusitis. Laryngoscope 117(7):1302–1306. doi:10.1097/MLG.0b013e31806009b0
Raad II, Darouiche RO, Hachem R, Abi-Said D, Safar H, Darnule T, Mansouri M, Morck D (1998) Antimicrobial durability and rare ultrastructural colonization of indwelling central catheters coated with minocycline and rifampin. Crit Care Med 26(2):219–224
Ravaioli S, Selan L, Visai L, Pirini V, Campoccia D, Maso A, Speziale P, Montanaro L, Arciola CR (2012) Staphylococcus lugdunensis, an aggressive coagulase-negative pathogen not to be underestimated. Int J Artif Organs 35(10):742–753. doi:10.5301/ijao.5000142
Richter AL, Gallagher KK (2016) Chronic invasive fungal sinusitis causing a pathologic Le Fort I fracture in an immunocompetent patient. Ear Nose Throat J 95(9):E1–E3
Rosenfeld RM, Piccirillo JF, Chandrasekhar SS, Brook I, Ashok Kumar K, Kramper M, Orlandi RR, Palmer JN, Patel ZM, Peters A, Walsh SA, Corrigan MD (2015) Clinical practice guideline (update): adult sinusitis. Otolaryngol Head Neck Surg 152(2 Suppl):S1–S39. doi:10.1177/0194599815572097
Saginur R, Stdenis M, Ferris W, Aaron SD, Chan F, Lee C, Ramotar K (2006) Multiple combination bactericidal testing of staphylococcal biofilms from implant-associated infections. Antimicrob Agents Chemother 50(1):55–61. doi:10.1128/aac.50.1.55-61.2006
Sanchez CJ Jr, Mende K, Beckius ML, Akers KS, Romano DR, Wenke JC, Murray CK (2013) Biofilm formation by clinical isolates and the implications in chronic infections. BMC Infect Dis 13:47. doi:10.1186/1471-2334-13-47
Shields RC, Mokhtar N, Ford M, Hall MJ, Burgess JG, ElBadawey MR, Jakubovics NS (2013) Efficacy of a marine bacterial nuclease against biofilm forming microorganisms isolated from chronic rhinosinusitis. PLoS One 8(2):e55339. doi:10.1371/journal.pone.0055339
Singhal D, Psaltis AJ, Foreman A, Wormald PJ (2010) The impact of biofilms on outcomes after endoscopic sinus surgery. Am J Rhinol Allergy 24(3):169–174. doi:10.2500/ajra.2010.24.3462
Snyman J, Claassen AJ, Botha PL (1988) A microbiological study of acute maxillary sinusitis in Bloemfontein. S Afr Med J 74(9):444–445
Sobin J, Engquist S, Nord CE (1992) Bacteriology of the maxillary sinus in healthy volunteers. Scand J Infect Dis 24(5):633–635
Speziale P, Pietrocola G, Foster TJ, Geoghegan JA (2014) Protein-based biofilm matrices in staphylococci. Front Cell Infect Microbiol 4:171. doi:10.3389/fcimb.2014.00171
Stepanovic S, Vukovic D, Hola V, Di Bonaventura G, Djukic S, Cirkovic I, Ruzicka F (2007) Quantification of biofilm in microtiter plates: overview of testing conditions and practical recommendations for assessment of biofilm production by staphylococci. APMIS 115(8):891–899. doi:10.1111/j.1600-0463.2007.apm_630.x
Sun F, Qu F, Ling Y, Mao P, Xia P, Chen H, Zhou D (2013) Biofilm-associated infections: antibiotic resistance and novel therapeutic strategies. Future Microbiol 8(7):877–886. doi:10.2217/fmb.13.58
Tote K, Vanden Berghe D, Maes L, Cos P (2008) A new colorimetric microtitre model for the detection of Staphylococcus aureus biofilms. Lett Appl Microbiol 46(2):249–254. doi:10.1111/j.1472-765X.2007.02298.x
Trampuz A, Zimmerli W (2006) Antimicrobial agents in orthopaedic surgery: prophylaxis and treatment. Drugs 66(8):1089–1105
Trampuz A, Piper KE, Jacobson MJ, Hanssen AD, Unni KK, Osmon DR, Mandrekar JN, Cockerill FR, Steckelberg JM, Greenleaf JF, Patel R (2007) Sonication of removed hip and knee prostheses for diagnosis of infection. N Engl J Med 357(7):654–663. doi:10.1056/NEJMoa061588
Vaudaux PE, Huggler E, Lerch PG, Morgenthaler JJ, Nydegger UE, Schumacher-Perdreau F, Lew PD, Waldvogel FA (1989) Inhibition by immunoglobulins of Staphylococcus aureus adherence to fibronectin-coated foreign surfaces. J Invest Surg 2(4):397–408
Vergara-Irigaray M, Valle J, Merino N, Latasa C, Garcia B, Ruiz de Los Mozos I, Solano C, Toledo-Arana A, Penades JR, Lasa I (2009) Relevant role of fibronectin-binding proteins in Staphylococcus aureus biofilm-associated foreign-body infections. Infect Immun 77(9):3978–3991. doi:10.1128/iai.00616-09
Wagner C, Aytac S, Hansch GM (2011) Biofilm growth on implants: bacteria prefer plasma coats. Int J Artif Organs 34(9):811–817. doi:10.5301/ijao.5000061
Walker JN, Horswill AR (2012) A coverslip-based technique for evaluating Staphylococcus aureus biofilm formation on human plasma. Front Cell Infect Microbiol 2:39. doi:10.3389/fcimb.2012.00039
Wecke J, Kersten T, Madela K, Moter A, Gobel UB, Friedmann A, Bernimoulin J (2000) A novel technique for monitoring the development of bacterial biofilms in human periodontal pockets. FEMS Microbiol Lett 191(1):95–101
Widmer AF, Gaechter A, Ochsner PE, Zimmerli W (1992) Antimicrobial treatment of orthopedic implant-related infections with rifampin combinations. Clin Infect Dis 14(6):1251–1253
Wood AJ, Fraser JD, Swift S, Patterson-Emanuelson EA, Amirapu S, Douglas RG (2012) Intramucosal bacterial microcolonies exist in chronic rhinosinusitis without inducing a local immune response. Am J Rhinol Allergy 26(4):265–270. doi:10.2500/ajra.2012.26.3779
Zheng Z, Stewart PS (2002) Penetration of rifampin through Staphylococcus epidermidis biofilms. Antimicrob Agents Chemother 46(3):900–903
Ziebuhr W, Krimmer V, Rachid S, Lossner I, Gotz F, Hacker J (1999) A novel mechanism of phase variation of virulence in Staphylococcus epidermidis: evidence for control of the polysaccharide intercellular adhesin synthesis by alternating insertion and excision of the insertion sequence element IS256. Mol Microbiol 32(2):345–356
Zimmerli W, Widmer AF, Blatter M, Frei R, Ochsner PE (1998) Role of rifampin for treatment of orthopedic implant-related staphylococcal infections: a randomized controlled trial. Foreign-body infection (FBI) study group. JAMA 279(19):1537–1541
Acknowledgments
Authors thank Dr. Ranieri Bizzarri for the helpful discussion related to the microscopy analysis. This work was supported by funds from University of Pisa and Consiglio Nazionale delle Ricerche.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2017 Springer International Publishing AG
About this chapter
Cite this chapter
Di Luca, M. et al. (2017). Detection of Biofilms in Biopsies from Chronic Rhinosinusitis Patients: In Vitro Biofilm Forming Ability and Antimicrobial Susceptibility Testing in Biofilm Mode of Growth of Isolated Bacteria. In: Donelli, G. (eds) Advances in Microbiology, Infectious Diseases and Public Health. Advances in Experimental Medicine and Biology(), vol 1057. Springer, Cham. https://doi.org/10.1007/5584_2017_34
Download citation
DOI: https://doi.org/10.1007/5584_2017_34
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-79016-9
Online ISBN: 978-3-319-79017-6
eBook Packages: Biomedical and Life SciencesBiomedical and Life Sciences (R0)