Abstract
Mitochondria are considered as the powerhouse of eukaryotic cells. They host several central metabolic processes fueling the oxidative phosphorylation pathway (OXPHOS) that produces ATP from its precursors ADP and inorganic phosphate Pi (PPi). The respiratory chain complexes responsible for the OXPHOS pathway are formed from complementary sets of protein subunits encoded by the nuclear genome and the mitochondrial genome, respectively. The expression of the mitochondrial genome requires a specific and fully active translation machinery from which aminoacyl-tRNA synthetases (aaRSs) are key actors. Whilst the macromolecules involved in mammalian mitochondrial translation have been under investigation for many years, there has been an explosion of interest in human mitochondrial aaRSs (mt-aaRSs) since the discovery of a large (and growing) number of mutations in these genes that are linked to a variety of neurodegenerative disorders. Herein we will review the present knowledge on mt-aaRSs in terms of their biogenesis, their connection to mitochondrial respiration, i.e., the respiratory chain (RC) complexes, and to the mitochondrial translation machinery. The pathology-related mutations detected so far are described, with special attention given to their impact on mt-aaRSs biogenesis, functioning, and/or subsequent activities. The collected data to date shed light on the diverse routes that are linking primary molecular possible impact of a mutation to its phenotypic expression. It is envisioned that a variety of mechanisms, inside and outside the translation machinery, would play a role on the heterogeneous manifestations of mitochondrial disorders.
Note: Rigorously, amino acid conversion of a given mutation should be preceded by the letter “p.” to indicate that the protein level is considered. For example, the 172C > G nucleotide change engenders the p.R58G mutation in DARS2 (referencing the gene) or mt-AspRS (referencing the protein). For sake of simplicity, the “p.” is omitted throughout the chapter.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
- Aminoacyl-tRNA synthetase
- Human mitochondrial disorders
- Pathology-related mutations
- Respiratory chain defects
1 Mt-aaRSs and Mitochondrial ATP Synthesis
In order to facilitate the understanding of the connection between human mt-aaRSs and ATP synthesis, the route from their place of encoding (the nucleus) to their place of biogenesis (the cytosol) and their place of use (the mitochondria) is schematized in Fig. 1.
From mitochondrial aminoacyl-tRNA synthetases to mitochondrial ATP synthesis. The route from the place of encoding of mt-aaRSs (the nucleus) to their place of biosynthesis (the cytosol) and their place of use (the mitochondria) is schematized. Mt-aaRSs biogenesis comprises mRNAs expression and processing (1), mt-aaRSs synthesis (2), import process into mitochondria (3a addressing; 3b translocation; 3c processing), and proper folding, oligomerization, and stability upon entry to mitochondria (4). Mt-aaRSs functioning includes amino acid activation, tRNA recognition, tRNA charging (5). Mt-aaRSs are devoted to the mitochondrial translation, and thus, to the synthesis of the 13 mt-DNA-encoded respiratory chain (RC) complexes (6), for which the activity (7) ultimately lead to ATP production (8). Of note, all other sub-units of the RC complexes (~70) are also imported from the cytosol
1.1 Mitochondrial Respiratory Chain Complexes
One of the most prominent functions of mitochondria is the production of cellular free energy in the form of ATP, in a process known as oxidative phosphorylation (OXPHOS). This process takes place in five large multi-subunit complexes (the respiratory chain complexes) that are located in the inner mitochondrial membrane (Fig. 2a). Complexes I to IV, accompanied by the mobile elements Coenzyme Q and cytochrome c, allow for the activity of Complex V, the ATP synthase. In mitochondria, the final oxidation of nutrients releases CO2 (mainly through the Krebs cycle) concomitantly with the reduction of NAD+ into NADH + H+ and FAD into FADH2. Oxidation of these hydrogen carriers involves the transfer of protons and electrons to the respiratory chain Complexes I and II respectively, followed by the channeling of these electrons through Complexes III and IV, where the electron is finally accepted by oxygen to form metabolic water. During electron transport, proton pumps in Complexes I, III, and IV become activated, leading to the expulsion of protons from the mitochondrial matrix to the mitochondrial intra-membrane space. The generated proton gradient (chemical potential) combined with the electron’s movement (electrical potential) lead to return of protons to the matrix, thus activating ATP synthase by a proton-motive force. This enzyme binds ADP to inorganic phosphate and generates ATP. In summary, final oxidation of nutrients into CO2 and H2O takes place inside mitochondria and is directly correlated to oxygen consumption and ATP synthesis by the respiratory chain complexes [1].
The respiratory chain complexes. (a) Organization of the complexes along the mitochondrial inner membrane. Complex I: NADH: ubiquinone reductase; Complex II: Succinate-coenzyme Q reductase; Complex III: Coenzyme Q: cytochrome c oxidoreductase; Complex IV: Cytochrome c oxidase; Complex V: ATP synthase. Q stands for Coenzyme Q, and cyt c for cytochrome c. Sub-units composition of the five complexes in given is Table 1. (b) Composition of the 13 mt-DNA-encoded sub-units
Respiratory chain complexes are large multi-protein complexes. Interestingly, the sets of proteins involved are of dual genetic origin. In humans, a total of 84 subunits and an additional 28 assembly factors are nuclear-encoded while 13 subunits are encoded by the mitochondrial genome (Table 1). These correspond to seven subunits of Complex I (NADH:ubiquinone reductase), one subunit of Complex III (Coenzyme Q: cytochrome c oxidoreductase), three subunits of Complex IV (cytochrome c oxidase), and two subunits of Complex V (ATP synthase). Complex II (succinate:coenzyme Q reductase) is the only complex formed exclusively by nuclear-encoded subunits. The size of the 13 mt-DNA-encoded proteins ranges from 68 amino acids (aa) (ATP8) to 603 aa (ND5) (Fig. 2b). The proteins are rather hydrophobic with 59.4% ± 8.5% aliphatic and aromatic residues, 29.8% ± 7.8% neutral residues, and 10.8% ± 2.6% charged residues. Leucine residues are present at the highest levels (14.4%); isoleucine, serine, and threonine are present at more than 7%, while some residues represent less than 3% (arginine, aspartate, cysteine, glutamine, glutamate, and lysine) of the protein compositions. It should be noted that a full and active set of mitochondrial translation components has been maintained for the synthesis of solely these 13 mt-DNA-encoded subunits.
1.2 The Human Mitochondrial Translation Machinery
The human mitochondrial genome is a circular double-stranded DNA of 16,569 bp [2]. This genome is tightly packed (with a single non-coding domain, the D-loop) and codes for the 13 respiratory chain subunits, in addition to 2 ribosomal RNAs (rRNAs) and 22 transfer RNAs (tRNAs). All three families of RNAs – mRNAs, rRNAs, and tRNAs – are processed from large primary transcripts according to the tRNA punctuation model [3]. The mitochondrial translation apparatus (Fig. 3) further involves a large number of proteins that are all nuclear-encoded, and are synthesized in the cytosol, before being imported into mitochondria for maturation. These include the full set of ribosomal proteins and ribosomal assembly proteins (translation initiation, elongation, termination factors) and tRNA maturation and modifying enzymes (enzymes cleaving the 5′- and 3′- ends of tRNAs primary transcripts, enzymes fixing the non-coded CCA 3′-end, enzymes of post-transcriptional modification). More than 100 proteins have been reported so far as being actors of the mitochondrial translation machinery [4–7] and will not be further discussed herein. Last but not least, a full set of nuclear-encoded aaRSs is required. These enzymes catalyze the specific esterification of tRNA 3′-ends with the corresponding amino acid so that the aminoacyl-tRNA (aa-tRNA) can be taken up by the translation factors and brought to the ribosome where the nascent protein is synthesized [8, 9]. The present knowledge of human mt-aaRSs will be discussed extensively below.
The mitochondrial translation machinery. The human mt-DNA codes for 13 respiratory chain subunits (mRNAs), 2 ribosomal RNAs (rRNAs), and 22 transfer RNAs (tRNAs). All other requested proteins, such as, e.g., ribosomal proteins and ribosomal assembly proteins, translation initiation, elongation, and termination factors, tRNA maturation and modifying enzymes, mt-aaRSs, are encoded by the nucleus, synthesized in the cytosol, and imported into the mitochondria
1.3 Link Between Mitochondrial Translation, Mitochondrial Respiration, and Mitochondrial Disorders
As mentioned above, ATP synthesis by mitochondria is dependent on the coordinated expression of nuclear and mitochondrial genes. First, there is a need to coordinate the biogenesis of the respiratory chain complexes along the inner mitochondrial membrane so that partner proteins find each other to form the individual multi-protein complexes. Second, there is also a need to coordinate the setup and maintenance of mitochondrial translation machinery. Above all, this involves efficient partnerships between mt-DNA-encoded RNAs and nuclear-encoded proteins, especially between rRNAs and ribosomal proteins to form active ribosomes and between tRNAs and aaRSs to allow for accurate synthesis of aa-tRNAs. Accordingly, there are key links between the aminoacylation activity of aaRSs in charge of the synthesis of the 13 mt-DNA-encoded proteins, and the activity of respiratory chain complexes. It can be anticipated that any dysfunction of a single macromolecule of the translation machinery may have severe impacts on the activity of the respiratory chain complexes.
Mitochondrial disorders were defined as pathologies with aberrant oxidative phosphorylation (OXPHOS). Potential causes include an aberrant ROS production, elevation of NADH/NAD+ ratio and lactate production, and/or ATP production deficiency. Defects were observed in a large variety of organs, and could manifest at any stage of life [10]. They were described in the late 1980s as exclusively related to mutations within the mt-DNA and thus maternally inherited. Additional disorders were subsequently associated with mutations within nuclear genes coding for proteins of mitochondrial location, and thus followed Mendelian inheritance. Mitochondrial disorders are nowadays classified according to the genetic origins of the involved-mutations. (1) The first category, actually the firstly reported, concerns mt-DNA-encoded RNAs. All the 22 mt-DNA-encoded tRNAs have been linked to pathology-related mutations. The most striking examples concern the tRNALys and tRNALeu, currently described as “hot spots” for mutations, and correlated respectively with Myoclonus Epilepsy with Ragged Red Fibers (MERRF [11]) and Mitochondrial Encephalomyopathy with Lactic Acidosis and Stroke-like episodes (MELAS [12]). More than 230 mutations in tRNA genes are presently referenced in the 2012 MITOMAP Human Mitochondrial Genome Database (http://www.mitomap.org). Mt-DNA also codes for 2 rRNAs, with around 50 disease-related mutations described that are most frequently connected with aminoglycoside-induced deafness or non-syndromic sensorineural deafness (DEAF). (2) The second category concerns mt-DNA-encoded proteins. They are all sub-units of the respiratory chain complexes, making those complexes sensitive to mt-DNA mutations. As an example, mutations in ND5 (subunit 5 of complex I) can lead to MELAS, Leigh syndrome, MERRF, or Leber’s Hereditary Optic Neuropathy (LHON) defects (reviewed in, e.g., [13]). (3) The third category, the most diverse, concerns nuclear-encoded proteins of mitochondrial location. On the one hand, mutations can affect proteins of the RC, which are directly contributing to OXPHOS (e.g., mutation in cytochrome oxidase subunits which are linked with Leigh syndrome, reviewed in [14]). On the other hand, mutations can affect proteins involved in the mt-DNA maintenance (e.g., DNA polymerase gamma, reviewed in [15]) and/or translation (e.g., mitochondrial elongation factor, reviewed in, e.g., [16]) interfering indirectly with OXPHOS. AaRSs of mitochondrial location belong to the last category. The recent discovery of mutations within mt-aaRS genes and the growing number of reported cases is opening a path to an emerging field of investigation (reviewed in, e.g., [6, 17]), and is the reason for a strong interest in the understanding of fundamental function of the mt-aaRSs and their implication in mitochondrial disorders.
Before reviewing in detail the molecular aspects of point mutations on mt-aaRSs properties, the various approaches available for the evaluation of respiratory chain complex activities used as tools either for diagnosis or for molecular investigation of this links between translation and respiration will be described.
1.4 Biochemical Analysis of Mitochondrial Respiration: A Potential Diagnostic Tool for the Detection of Mitochondrial Translation Defects
Because mitochondria provide much of the cellular energy, mitochondrial disorders preferentially affect tissues with high energy demands, including brain, muscle, heart, and endocrine systems. Consequently, mitochondrial defects play a central role in hereditary mitochondrial diseases, ischemia reperfusion injury, heart failure, metabolic syndrome, neurodegenerative diseases, and cancer [18, 19]. There are remarkably diverse causes for mitochondrial disorders. These may be linked to the dual genetic systems encoding components of the respiratory chain complexes, to the need for mitochondrial translation machinery, but also to mechanisms required for the biosynthesis and maintenance of mt-DNA and to the biogenesis of the organelle itself. Moreover, each cell may contain hundreds to thousands of copies of the mitochondrial genome. The distribution of the affected tissues and the proportion of mutant to wild-type mt-DNA (termed heteroplasmy) lead to clinical manifestations, which are remarkably variable and heterogeneous. An additional breakthrough came from the later discovery of mutations within nuclear genes as the causes for similar diseases. Therefore, the establishment of a mitochondrial disorder diagnosis can be very difficult. It requires an evaluation of the family pedigree, in conjunction with a thorough assessment of the clinical, imaging, and muscle biopsy analysis [20]. Isolated OXPHOS deficiencies are generally caused by mutations in genes encoding subunits of the OXPHOS system. Combined deficiencies in the respiratory chain complexes may reflect the consequence of mutations in mt-DNA-encoded tRNAs or rRNAs, or are due to rearrangements or depletion of mt-DNA [21]. They may also reflect a dysfunction of the mitochondrial translation machinery.
Table 2 summarizes some of the various experimental procedures used to evaluate the impact of a mutation on the different steps from mt-aaRS expression to mitochondrial ATP synthesis. It includes the initial screening procedure of muscle biopsy analysis, i.e., histochemistry and immunohistochemistry. The activity of the five multiprotein enzymatic complexes can be assayed globally by measuring the mitochondrial inner membrane electrochemical potential, oxygen consumption, or ATP synthesis, or assayed individually by measurement of their enzymatic activities. This is performed by polarography. The structural integrity of the multiprotein complexes, evaluated by blue native gel electrophoresis is also increasingly used for diagnostics [22]. Finally, once the diagnosis is performed, it is possible to evaluate the impact of mutations on the rate of mitochondrial protein synthesis using chase experiments on specific recombinant cell-lines [23].
Several histochemical and immunohistochemical methods can be used as reliable morphological tools in order to visualize respiratory chain abnormalities on tissue sections. Classical histochemistry techniques allow visualizing succinate dehydrogenase (SDH) and cytochrome c oxidase (COX) activity. Indeed, the most informative histochemical impairment of mitochondria in skeletal muscle is ragged red fibers (RRF), observed on frozen sections traditionally with the modified Gomori trichrome method [24]. Since the accumulation of material other than mitochondria sometimes simulates a RRF appearance, the identification of deposits suspected of being mitochondrial proliferation should be confirmed by histochemical staining of oxidative enzymes as SDH and COX. Muscle from mitochondrial myopathy associated with mt-DNA mutations tends to show a mosaic expression of COX consisting of a variable number of COX-deficient and COX-positive fibers, and RRFs can be COX negative or COX positive. The mosaic pattern of COX expression could be considered as the histochemical signature of a heteroplasmic mt-DNA mutation affecting the expression of mt-DNA-encoded genes in skeletal muscle [25]. Immunohistochemical methods allow the visualization of the expression of several mt-DNA and nuclear-encoded subunits of the respiratory chain. An antibody against COX IV is routinely used as a probe for nuclear-encoded mitochondrial protein and an antibody against COX II as a probe for mt-DNA-encoded protein. However, any other combination of antibodies can also be used.
Polarography (spectrophotometric assays) can be applied to both tissue samples and cultured cells and is designed to assess the enzymatic activity of the individual OXPHOS Complexes I–V, along with the Krebs cycle enzyme citrate synthase as a mitochondrial control. Determining the enzymatic activities can be valuable in defining isolated or multicomplex disorders and may be relevant to the design of future molecular investigations [26]. Different assays have to be performed in order to analyze each complex separately. Assays for Complexes I, II, II + III, III, and IV are routinely performed when there is a suspicion of mitochondrial defects. The principle of polarographic approach is based on the fact that mitochondria require oxygen to produce ATP. The rate of oxygen consumption from isolated mitochondria or directly in skinned fibers is a useful and valuable technique in the research and evaluation of mitochondrial dysfunction and disease, because ADP-dependent oxygen consumption directly reflects OXPHOS efficiency [27–31]. In the presence of oxidizable substrates, freshly isolated mitochondria are introduced into the polarograph and oxygen consumption is measured in the presence of exogenously added ADP as well as several inhibitors. The typical parameters determined from mitochondrial polarography include state III rate (maximal mitochondrial respiration with ADP), state IV rate (basal mitochondrial respiration), and RCR (respiratory control ratio or state III rate/state IV rate). RCR is a good indicator of the integrity of the inner membrane of the isolated mitochondria and is sensitive for indicating OXPHOS defects. The ratio of ADP consumed/oxygen consumed during the experiment is a direct reflection of phosphorylation efficiency and can indicate abnormalities of the ATP synthase activity or uncoupling between the activities of Complexes I to IV and complex V (ATP synthase). It is possible to explore the respiratory parameters of skeletal muscle with permeabilized muscle fibers, thus skipping a mitochondria purification step [32]. Muscle fibers are permeabilized by saponin, allowing respiratory substrates and inhibitors to reach the mitochondria [31, 33].
Blue native polyacrylamide (or agarose) gel electrophoresis (BN-PAGE) allows for the isolation of intact respiratory chain complexes and analysis of their subunit content (reviewed in [34]). Briefly, after solubilization of mitochondria in the presence of dodecylmaltoside, large complexes are first separated by native gels electrophoresis on low percentage polyacrylamide or agarose gel. A second dimension electrophoresis is then performed under non-native conditions in the presence of SDS and β-mercaptoethanol using a 10% polyacrylamide gel. Individual subunits are detected by western blotting or mass spectrometry. The two-dimensional separation approach can also be adapted to perform in gel activity assays to address the dynamics of protein synthesis and complex assembly (in combination with pulse-chase labeling of proteins in cultured cells).
In conclusion, a variety of biochemical approaches are available to evaluate mitochondrial function. These form a powerful toolkit that can be used to diagnose the mitochondrial origin of a disorder. However, the gap is large between the dysfunction of the respiratory chain (as a whole, or as individual complexes) and the understanding of the dysfunction at the molecular level. Indeed, considering the translation rate of each of the 13 mt-DNA-encoded proteins as the sole outcome resulting from defects in tRNAs and/or aaRSs is too simplistic. The links between the mitochondrial translation of a given aaRS on the one hand, and ATP production on the other, involve a number of issues that need to be explored. Some of these will be discussed in what follows. One must also take into account the possibility of alternative functions of mt-aaRSs, outside their strict housekeeping role in translation.
2 Mt-aaRSs in Mitochondrial Translation
The housekeeping function of aaRSs is to provide aminoacylated-tRNAs (aa-tRNAs) for translation. Enzymes connect tRNAs with their corresponding amino acid in an efficient and specific way through a two-step reaction named aminoacylation. In the first step, the amino acid is activated by ATP into an aminoacyl-adenylate (followed by the release of PPi). In the second step, the activated amino acid is transferred onto the cognate tRNA, releasing AMP. The formation of the 20 canonical aa-tRNA species in human mitochondria requires the import of a complete set of mt-targeted aaRSs encoded by the nuclear genome. The faster evolution rate of mt-DNA than the nuclear genome [35, 36] leads to abnormal RNAs, shrunken in size and often lacking important signals. For instance, most mt-tRNAs have shortened sizes, miss crucial folding and recognition nucleotides as compared to “classical” tRNAs, and are more flexible [6, 7, 37, 38]. Mechanisms compensating for the degeneration of mt-tRNAs remain mainly unsolved and raise the question of molecular adaptation of partner proteins, especially mt-aaRSs. The structural and functional deciphering of the set of aaRSs of human mitochondrial location is at the early stages.
2.1 Nuclear-Encoded aaRSs of Mitochondrial Location and Evolutionary Considerations
The nuclear gene annotation of the human aaRSs of mitochondrial location was completed a decade ago [8]. Access to gene sequences highlighted that the set of aaRSs dedicated to translation in human mitochondria is mainly different from the set acting in the cytosolic translation (Fig. 4). This concerns 17 out of the 19 pairs of aaRS. The two exceptions concern the GlyRSs and LysRSs. GlyRSs are generated from two translation initiation sites on the same gene, leading to one enzyme with a mitochondrial targeting sequence (MTS), mt-GlyRS, and a second without, cytosolic GlyRS [39, 40]. With the LysRSs, an alternative mRNA splicing pathway allows for the insertion – or not – of the nucleotide sequence coding for the MTS, leading to two mature mt- and cytosolic LysRSs differing only by a few residues at their N-terminus [41]. No gene coding for mt-GlnRS, the 20th synthetase, has been found so far, leaving open the question about how glutaminylation in human mitochondria is performed. Among possible explanations, it is proposed that either the sequence of human mt-GlnRS has evolved so much that it has become unrelated to any of the known GlnRSs, or its function is fulfilled by an mt-addressed version of cyt-GlnRS (as proposed in yeast [42]), or that the synthesis of mt Gln-tRNAGln occurs via an indirect pathway (the transamidation pathway) involving misacylation of tRNAGln by a non-discriminative GluRS followed by Glu-amidation [43]. The existence of an indirect pathway in mitochondria was demonstrated in the cases of plants [44, 45], yeast [46], and more recently humans [47]. However, the coexistence of direct and indirect pathways for Gln-tRNAGln synthesis in yeast and mammalian mitochondria is still under consideration [48, 49].
Two sets of genes for human cytosolic and mitochondrial aaRSs. Achievement of human genome sequencing and gene annotation of the human aaRSs of mitochondrial location [8] reveal the presence of two distinct sets of nuclear genes for aaRSs (with two exceptions, see text). One set (in red) codes for the aaRSs of cytosolic location. The second set (in blue) codes for the aaRSs of mitochondrial location. The latter distinguishes by the presence of an encoded N-terminal mitochondrial targeting sequence (MTS, in yellow). The two sets of genes are translated via the cytosolic translation machinery. Sequences that are subsequently addressed to mitochondria are maintained unfolded in the cytosol, pending their entry into the organelle
Despite the conventional view of the endosymbiotic origin of mitochondria [50], the source of nuclear genes for mt-addressed aaRSs is diverse and not necessarily easy to trace back. Some of the mt-addressed aaRSs originate from the bacterial domain, but none specifically from the alpha-proteobacteria, although the alpha-proteobacterial contribution to the mitochondrial genome is well established. This favors the hypothesis that mt-aaRSs have been acquired by numerous post-endosymbiotic and/or lateral gene transfer events, from sources representative of all kingdoms of life [51]. The precise knowledge on the origin of all human mt-aaRS genes is necessary for the investigation of pathology-related mutations (see below) and will be of help in building up homology models in cases where crystallographic structures for mt-aaRS are not available. This global view on the origin of all human mt-aaRS genes will be established soon (Sissler et al., in preparation).
2.2 Mt-aaRSs are Imported Proteins
Human mt-aaRSs are translated within the cytosol and subsequently imported into the mitochondria thanks to the presence of MTSs. As is the case for the vast majority of proteins of mitochondrial matrix and/or inner membrane location, these sequences are predicted to be located at the N-termini of precursor mt-aaRSs [8]. However, to date, no consensus sequences have been deciphered. An MTS typically consists of ~15–50 amino acids including numerous positively charged residues (e.g., lysines and arginines), and forms amphipathic alpha-helices [52]. The MTS first directs the precursor proteins to mitochondria where they are further translocated by the translocase complexes of the outer (TOM) and inner (TIM) membranes (reviewed in, e.g., [53–59]). Upon arrival in the matrix, MTS are proteolytically cleaved by the mitochondrial processing peptidase (MPP). This process was recently shown possibly to affect the half-life of the proteins [60]. Removal of the pre-sequence exposes new amino-termini of the imported proteins, which may contain a stabilizing or a destabilizing amino acid (bulky hydrophobic residues are typically destabilizing). The N-end rule indeed states that regulation of the proteolytic degradation is closely related to the N-terminal residue of proteins [61]. It has recently been proposed that two additional peptidases (that function subsequently to MPP) are implicated in protein stabilization by removing the newly exposed N-terminal destabilizing residue(s). The first is the intermediate cleaving peptidase Icp55, which removes a single amino acid. The second is the mitochondrial intermediate peptidase Oct1, which removes an octapeptide. Accordingly, the processing of imported proteins is closely connected to protein turnover and quality control (reviewed in [54]).
Cleavage (maturation) sites of the MTS for human mt-aaRSs are so far mostly defined according to theoretical predictions based on computer programs (e.g., Predotar http://urgi.versailles.inra.fr/predotar/predotar.html, MitoProt http://ihg.gsf.de/ihg/mitoprot.html, TargetP http://www.cbs.dtu.dk/services/TargetP/, and iPSORT http://ipsort.hgc.jp/). However, the expression of recombinant human mt-aaRS proteins, deprived of theoretically predicted MTSs, appears to be difficult due to low solubility and the tendency of proteins to aggregate [9, 62]. This is likely to indicate that many predictions may be incorrect. Indeed, inaccurate prediction of the cleavage site was previously shown to be responsible for suboptimal expression of human mt-LeuRS. Only the LeuRS variant deprived of its 39 N-terminal amino acids was sufficiently overexpressed in Escherichia coli, efficiently purified, and fully active, while the variant deprived of the predicted 21 amino acids remained insoluble [63, 64]. Along the same line, the re-design of the N-terminus of human mt-AspRS enhances expression, solubility, and crystallizability of the mitochondrial protein [65, 66]. Discrepancies are also apparent between the predicted cleavages sites, the starting amino acid of the recombinant proteins, and the first residue visible in the established crystallographic structures to date (reviewed in [65]).
None of the above-mentioned examples have the experimental exact cleavage points of the mature proteins been established so far. These examples however emphasized that the preparation of stable recombinant molecules would gain from optimized criteria to predict MTS cleavage sites. A more systematic effort to determine experimentally the precise N-terminus of mature mitochondrial proteins (as done for, e.g., the yeast mt proteome [60]) would be of help to determine unambiguously the sequence of a functional mt protein. However, this analysis remains difficult to perform experimentally (by sequencing or mass spectrometry) due to the minute amount of protein that can be isolated from human mitochondria and because of the risk of secondary proteolysis.
2.3 Structural Insights of mt-aaRSs
When considering their primary structures, all mt-aaRSs fall into the expected classes of the aaRSs as originally defined in [67, 68], with signature motifs being respectively HIGH and KMSK for class I enzymes, and motifs 1, 2, and 3 for class II enzymes. Striking divergences are, however, observed when considering their modular organizations. Modular design of the aaRSs is a result of a patchwork assembly of different functional modules during evolution. Minimal cores are the catalytic domain and the tRNA anticodon binding domain that are possibly surrounded by additional components for structural or functional purposes. Those are either remnants from early ancestors or structural inventions for functional expansion (e.g., [69]). The most striking observation concerns the divergence between the tetrameric cytosolic PheRS (α2β2) and the monomeric (α) version found in human mitochondria [70], a situation also observed for PheRSs in yeast [71]. Human mt-PheRS possesses the minimum set of structural domains, making this enzyme the smallest exhibiting aminoacylation activity and the only class II monomeric synthetase [72]. Another striking observation is that two independent coding sequences have been found for mt-GluRS and mt-ProRS, as opposed to a single gene for both activities in the human cytosol leading to the bifunctional GluProRS [73].
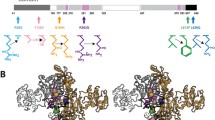
Although the first crystallographic structure of a bacterial aaRS was published three decades ago (TyrRS from Bacillus stearothermophilus [74]), the first 3D structures of mt enzymes were only established recently: a bovine structure in 2005 (mt-SerRS [75]), the first human structure in 2007 (mt-TyrRS [76]), followed by two additional ones in 2012 (mt-PheRS [72] and mt-AspRS [66]) (Fig. 5). The time lag between resolving structures of bacterial aaRS and mammalian mt-aaRSs was mainly due to the difficulties involved in producing large amounts of stable mitochondrial proteins (defining the N-terminus leading to a soluble protein). Resolution of crystallographic structures and investigation of biophysical properties of mt-aaRSs reveal similarities, but also distinctive features, when compared to related prokaryotic homologs (the four mt-aaRSs for which crystallographic structures have been obtained are of prokaryotic origin [51]).
Known crystallographic structures of mammalian mt-aaRSs. (A) Bovine mt-SerRS [75], where the specific “distal helix” and “C-tail” are emphasized in green and red, respectively. In addition, the bacterial-type N-terminal long α-helices hairpin arm is shown in orange. (B) Human mt-AspRS [66], where the bacterial insertion domain is highlighted in light green. (C) Human mt-TyrRS [76], where the CP1 and the α-helical domains are indicated in red and gray, respectively. Note that the S4-like domain is missing in this structure. (D) Human mt-PheRS in the –closed– state [77], and in the –open– state, complexed within Thermus thermophilus tRNAPhe (in white) [72]. Binding of tRNA engenders a drastic conformational change of mt-PheRS through ~160° hinge-type rotation and the global repositioning of the anticodon binding domain. For all structures the catalytic core is in yellow, the anticodon binding domain in orange, and the hinge region in black. When appropriate, the second dimer is displayed in light gray
Structural idiosyncrasies (Fig. 5) concern, for instance, the very unique structural organization of mt-PheRS (monomer instead of heterotetramer [77]) or the presence of two distinctive insertions in mt-SerRS (an 8 aa amino-terminal “distal helix” and a carboxy-terminal “C-tail” composed of an over 40 Å long flexible loop stretching away from the body of the monomer [75]). More generally, it has been observed that, besides having similar architectures to prokaryotic homologs, mitochondrial enzymes are distinguished by more electropositive surface potentials (specifically along the tRNA binding surface) or by enlarged grooves for tRNA accommodation. The latest aspect is visible for instance in mt-AspRS, where the angle formed by the bacterial-specific insertion domain and the catalytic domain is more opened by 26°C [66]. This can also be seen in mt-PheRS where the PheRS/tRNAPhe complex formation was shown to be accompanied by a considerable rearrangement through an ~160° hinge-type rotation from a –closed– to an –open– state of the PheRS and the global repositioning of the anticodon binding domain upon tRNA binding [72, 77]. In addition, an alternative interaction network has been observed at the subunit interface of the dimeric mt-AspRS (weaker in terms of salt-bridges and hydrogen bonds). Biophysical investigations also demonstrated a thermal stability reduced by as much as 12°C for mt-AspRS, compared to E. coli AspRS [66]. Finally, a gain of plasticity is proposed for both the mt-TyrRS, where the KMSKS loop is rather remote from the active site, explaining the relative lacks of constraints in the structure [76], and the mt-AspRS, where unusual thermodynamic properties of tRNA binding are observed [66]. It has been suggested that the gain of plasticity may be a more general property of mt-aaRSs, as they have to deal with degenerated mt-tRNAs [66].
2.4 Some Functional Peculiarities
The bacterial origin (established for many of the mt-aaRSs) predicts that most of the mt-aaRSs should behave as prokaryotic aaRSs. However, this is not the case, raising interesting questions regarding the evolution of macromolecules of the mitochondrial translation machinery. The human mt-AspRS has been extensively studied along these lines. It shares 43% of identical residues (including residues specific for all AspRSs), the same modular organization (including the bacterial-type insertion and C-terminal extension domains), and the same architecture as E. coli AspRS, a representative bacterial homolog [66]. However, and despite the fact that the two enzymes are likely descendants from a common ancestor, numerous functional idiosyncrasies/discrepancies were reported. Indeed, the mt-AspRS exhibits a reduced catalytic efficiency [8, 9], requires a minimal set of recognition determinants within its cognate tRNA [78], displays a higher sensitivity to small substrate analogs [79], is able to cross aminoacylate bacterial tRNAs (while the bacterial enzyme unilaterally recognizes bacterial tRNAs [80, 81]), and shows an increased intrinsic plasticity when compared to its bacterial homolog [66].
It is proposed that all structural and functional peculiarities of the mt-aaRSs (exemplified here by the mt-AspRS) with respect to the bacterial homologs may represent an evolutionary process, allowing nuclear-encoded proteins to cooperate with degenerated organelle RNAs [66].
3 Mt-aaRSs in Human Disorders
As already evoked, a breakthrough took place in 2007 with the discovery of a first set of mutations present in the nuclear gene of an mt-aaRS, namely mt-AspRS [82]. This first discovery was followed very rapidly by the description of numerous additional mutations not only on the same gene but also on other mt-aaRS-coding genes, so that half of them are presently known to be affected [6, 17]. This discovery sheds light on a new family of nuclear genes involved in human disorders allowing the new naming of “mt-aaRS disorders.”
3.1 Discovery of aaRS-Related Disorders
Mutations in DARS2, the nuclear gene coding for mt-AspRS, were first found in 2007 in patients with cerebral white matter abnormalities of unknown origin [82]. These abnormalities were part of a childhood-onset disorder called Leukoencephalopathy with Brain stem and Spinal cord involvement and Lactate elevation (LBSL). Since this first discovery, mutations in eight additional mt-aaRS-encoding genes have been reported (Table 3). They hit RARS2 (patients with PontoCerebellar Hypoplasia type 6, PCH6), YARS2 (Myopathy, Lactic Acidosis and Sideroblastic Anemia, MLASA syndrome), SARS2 (HyperUricemia, Pulmonary hypertensions and Renal failure in infancy and Alkalosis, HUPRA syndrome), HARS2 (Perrault Syndrome, PS), AARS2 (Infantile Mitochondrial Cardiomyopathy), MARS2 (Autosomal Recessive Spastic Ataxia with Leukoencephalopathy, ARSAL), EARS2 (Early-onset Leukoencephalopathy with Thalamus and Brainstem involvement and High Lactate), and FARS2 (Infantile mitochondrial Alpers Encephalopathy). An observation among the numerous reported cases is that despite a dominant effect on brain and neuronal system, sporadic manifestations also occur in skeletal muscle, kidney, lung, and/or heart.
3.2 The Present-Day List of Pathology-Related Mutations Within mt-aaRS Encoding Genes
Nine mt-aaRS genes are currently known to harbor a total of 65 mutations, found in patients in 64 different genetic combinations (Table 4). All mutations can also be visualized on schematic representations of modular organizations of the proteins (Fig. 6). The most prominent affected gene is DARS2, located on chromosome I. It comprises 32,475 bp, codes for 17 exons and is translated into a 645 aa long mt-AspRS. To date, 28 different mutations are known: 8 nonsense mutations (frameshift, premature stop), 16 missense mutations (amino acid exchange), and 4 insertions/deletions have been described in 13 different reports. These mutations are distributed over nearly all exons and, thus, are found in all protein domains, including the predicted MTS (S45G) and the hinge region (C152F and M134_K165del). In addition to these mutations found on one allele of the gene, almost all LBSL patients have a mutation in a polypyrimidine tract at the 3′-end of intron 2, which is found on the second allele. This mutation affects correct splicing of the third exon, which leads to a frameshift and a premature stop (R76SfsX5). This frameshift mutation is “leaky,” leading to a decrease but not zero expression of full-length mt-AspRS. Beside these typically compound heterozygous states of LBSL patients, homozygous DARS2 mutations have been described more recently. Homozygous patients harbor either the R76SfsX5 mutation or the R609W mutation (in the bacterial insertion domain).
The EARS2 gene codes for mt-GluRS and is located on chromosome XVI. It comprises 32,980 bp, nine exons and encodes a 523 aa long protein. Today, 16 different mutations are known: 3 nonsenses, 12 missenses, and 1 insertion. The catalytic domain is dominantly affected. The two mutations (R7X and M1X) localized in the predicted MTS are nonsense mutation and certainly lead to truncated translation products. Among those, only one mutation (K65E) is in a homozygous state. The gene RARS2 codes for mt-ArgRS, is located on chromosome VI, consists of 75,618 bp, 20 exons, and codes for a 562 aa long protein. Mutations in this gene affect all parts of the protein except the dinucleotide-binding domain. Ten different mutations are reported: a single nonsense (Q12fsX25), seven missenses, and two deletions (K158del, R504_L528del). In addition, three homozygous patients were found harboring a combination of two silent mutations (K568K and K291R) with one additional mutation (IVS2 + 5) causing exon 2 skipping. The patient’s major transcript lacked exon 2, but a faint, normal-sized fragment was also seen. The gene FARS2 is 403,026 bp long, codes for mt-PheRS, and is located on chromosome VI. It has six exons and codes for a 451 aa long protein. Today four different missense mutations have been reported. Just the Y144C one (in the anticodon binding domain) is in a homozygote state. The gene HARS2 codes for mt-HisRS, is located on chromosome V, and consists of 6,900 bp and 13 exons. The corresponding protein is 506 aa long. There is presently only one reported case in which patients are compound heterozygotes. The mutation on one allele is a missense (V368L). The mutation on the second allele produces either a missense replacement (L200V) or creates an additional splice site, inducing the deletion of 11 aa (L200_K211del). The YARS2 gene, coding for mt-TyrRS, is localized on chromosome XII, is 8,630 bp long, harbors five exons, and codes for a 477 aa long protein. For this aaRSs, only two homozygote mutations are known, both being localized in the catalytic domain. The AARS2 gene codes for mt-AlaRS, is 13,672 bp long, localized on chromosome VI, has 22 exons, and codes for a 985 aa long protein. There are presently two-reported cases in which patients are harboring missense mutations (either homozygote or compound heterozygote), all localized in the catalytic domain. The SARS2 gene codes for mt-SerRS, is located on chromosome XIX, is 15,127 bp long, and contains 16 exons. Only one homozygote mutation is reported, localized in the catalytic domain of the 518 aa long protein. It should be noted that the MARS2 gene, located on chromosome II, is composed of just one exon of 1,779 bp length. It codes for a 593 aa long protein, named mt-MetRS. Interestingly, no “classical” mutation has been reported for this gene but complex rearrangements were shown to be the cause of ARSAL. The single-exon composition of the gene permits duplication events of either the full exon or part of it, leading to the homozygous state or compound heterozygous state of the patients. An additional situation with a large insertion in one of the alleles has also been reported.
In summary, none of the chromosomes is a “hot spot” for pathology-related mutations affecting mt-aaRSs, and neither exons nor protein domains have obvious favored mutation sites. Presently, DARS2 is the most frequently hit gene. However this may not indicate a peculiar mutational exposure of this gene, but is more likely to be due to intensive investigations of this firstly reported example of an mt-aaRS gene correlated with a disease.
3.3 Compound Heterozygous vs. Homozygous States
Among the 64 reported combinations of mutations, 53 are compound heterozygous and 11 are homozygous (excluding the puzzling combinations found for MARS2; see below). In all cases, the parents are unaffected heterozygous carriers of one mutation, leading to autosomal recessive mutations that affect the two alleles in the children. Figure 7 schematically summarizes all observed situations that are combining splicing, missense, nonsense, deletion, and rearrangements defects.
From the gene to the protein: impact of pathology related mutations. The schematic views illustrate reported combinations of compound heterozygous and homozygous mutations (red crosses), and gene rearrangements on a genetic level, as well as possible impacts on corresponding proteins. Exonic organization is represented here in a simplified way (gray boxes schematize exons). Truncated proteins result from either nonsense mutations or splicing defects (reported to be “leaky” so that a reduced amount of full-length protein remains expressed, as shown by the thinner bars). The natural oligomeric status of mt-aaRSs is recalled on the right. nd stands for not determined
Compound heterozygous status is dominantly observed. In most of the cases, mutations in the first allele produce a splicing defect, resulting in reduced expression of the protein. The second allele carries a missense mutation. Such combinations were reported for DARS2 [82–85] and RARS2 [86, 87]. In both situations the splicing defect is “leaky,” so that a small amount of wild-type protein remains expressed, which is likely to be sufficient to support basal aminoacylation activity. The residual expression of wild-type protein is mandatory in the situation where the splicing defect is combined with nonsense mutations (which completely abolishes protein expression) as was reported for, e.g., DARS2 (R76SfsX5/Q357X [88]) and RARS2 (Q12fsX25/L13RfsX15 [86]). Other examples of nonsense mutations are found, but combined with missense mutations of likely moderate consequences. This is, for instance, reported for EARS2 (R7X or Y398X combined with R108W [89]). Another possible impact of mutation is the deletion of one or more amino acids within the protein. Mt-HisRS harboring the L200_K211 deletion has been suggested to be unstable when mutants are transiently expressed in human cells [90]. Deletions have also been reported in RARS2 and DARS2 (combined in the latest with the abundant R76SfsX5 mutation). However, their possible impacts on protein expression level or stability remains unclear [82, 83, 91, 92]. As a last example, combinations of two different missense mutations have been reported for EARS2, FARS2, RARS2, DARS2, and AARS2. In these cases, it is assumed that proteins are expressed but folding, structure, stability, and/or activity could be affected (see below).
Figure 7 recalls the natural oligomeric status of mt-aaRSs: some are monomers (mt-PheRS, mt-GluRS, mt-ArgRS), some are dimers (mt-AspRS, mt-SerRS, mt-TyRS, mt-HisRS, mt-MetRS), and one (mt-AlaRS) is homotetramer. A consequence of heterozygosity is the production of two distinct polypeptide chains that can randomly associate to build a dimer (or a tetramer), which theoretically leads to an equal proportion between the two possible homodimers (each harboring the same mutation) and the heterodimer (where each constitutive monomer is harboring a different mutation). However, it has been reported that some of the mutations have an impact on the oligomerization rate, leaving out the random association of mutated polypeptide chains. For instance, mt-HisRS, having the V368L mutation, oligomerizes more efficiently than any combinations, including the wild-type polypeptide [90].
The discovery of homozygous mutations was quite unexpected. In fact, mutations in DARS2 were found initially only in a compound heterozygous state, suggesting that the activity of mutant mt-AspRS homodimers may be incompatible with life. It was then proposed that dimers carrying two different mutations would have more residual functional activity than those carrying the same mutation, and thus would not yield to a lethal phenotype [93]. The recent discovery of homozygous mutations of DARS2 correlated with LBSL in a German patient and in a Japanese family [94, 95] ruled out this conviction. Therefore, the sole possibility for homozygous mutations to be compatible with the survival of the patient is that the mutation does not exert a too severe effect. As an example, the homozygous R76fsX5 mutation in DARS2 from an LBSL patient induces a splicing defect that was demonstrated to be “leaky.” This allows the expression of a small amount of wild-type protein likely to be sufficient to support some basal activity [96]. Today, homozygous missense mutations are found in AARS2, DARS2, EARS2, FARS2, RARS2, YARS2, and SARS2. Their “moderate” effect is not obvious since none of the observed-mutations (e.g., K65E in EARS2, Y144C in FARS2, D390G in SARS2, or F52L and G46D in YARS2) conserve any of the physico-chemical properties of the amino acids (isostericity, net charge, hydrophobicity, …). It has however been demonstrated that mt-TyrRS carrying the F52L mutation remains catalytically active, with only a twofold reduced aminoacylation rate [97], emphasizing that volume/polarity of the amino acid cannot be the only parameter to take into account. Possible neighborhood effect and structural impacts are discussed below.
In this discussion, MARS2 is an exception. No “classical” missense or nonsense mutations are observed, but instead complex gene rearrangements have been reported. As already mentioned, this gene is composed of a single exon. A local genomic instability and/or recombination errors have been hypothesized to be the cause of template switching during DNA replication [98]. As a consequence, duplication events of either the full exon or part of it are observed in patients, leading to either homozygous or compound heterozygous states. An additional situation with a large insertion of 300 bp in one of the alleles has also been reported.
4 Diverse Molecular Impacts
A mutation in an mt-aaRS gene can have numerous molecular consequences, affecting either the biogenesis of the enzyme itself, and/or its import and maturation within the organelle, and/or its functional properties. Figure 1 (Part 1) schematized the link of these different steps with ATP synthesis. Biogenesis of mt-aaRSs involves expression and processing of the corresponding mRNA, protein expression, and stability in the cytosol and addressing to mitochondria. Importing into mitochondria requires several steps. It is followed by maturation steps of imported proteins upon entry into mitochondria. Housekeeping function of mature mt-aaRS corresponds to amino acid activation, tRNA recognition, and tRNA aminoacylation. Pathology-related mutations may thus have a direct effect on the mitochondrial translation machinery by impacting mt-aaRS biogenesis, localization (see Sect. 4.1) and/or function (see Sect. 4.2). As a consequence, the translation efficiency/rate of the full set of (or specific) mt-DNA-encoded subunits of respiratory chain complexes may be affected as well, impacting respiratory chain complexes activities and, finally, ATP synthesis (see Sect. 4.3).
4.1 Impact of Mutations on mt-aaRSs Biogenesis
4.1.1 Defects in mt-aaRS mRNAs Processing and Expression
The previously mentioned complex gene rearrangement of MARS2 leads to an increased amount of transcript (due to duplication of full size gene and/or of regulatory elements). However, no increase in the amount of proteins is observed. It was suggested that corresponding mRNAs undergo transcriptional regulatory event(s) drastically lowering mRNA stability and thus leading to a reduction of 40–80% of the normal protein level [98]. In DARS2 and RARS2, several intronic mutations were reported to affect pre-mRNAs processing. Most of the LBSL patients have mutations (of different types) within intron 2, which affects the correct splicing of exon 3. Similarly, some PCH6 patients have an IVS2 + 5(A > G) mutation, which leads to exon 2 skipping. Exclusion of exon 3 (in DARS2) or of exon 2 (in RARS2) causes a frameshift (since the two exons are asymmetric) and generates a premature stop codon. In the two situations, it is speculated that the leaky nature of the splicing defect allows for the synthesis of a small (sufficient) amount of wild-type mRNA in most tissues [94, 96, 99, 100]. However, the selective vulnerability within the nervous system is explained by tissue-specific differences in the concentration of the splicing factors (reduced in neural cell) and the presence of rather weak splice sites. In agreement, 5′-splice site of exon 3 in DARS2 and 3′-splice site of exon 2 in RARS2 were shown to deviate markedly from the consensus and to have low splicing scores (even in the absence of disease-causing mutations). As a consequence, the exclusion of related exons induced by the mutations is augmented in neural cells [96, 101]. Another example of mis-splicing concerns mutation Q12R(Q12fsX25) in RARS2. This mutation interferes with a splicing-enhancer element and causes the retention of 221 bp from intron 1, a consequent frameshift, and the truncation of protein after residue 25 [87].
4.1.2 Defects in mt-aaRSs Expression and Stability
A defect in protein expression and stability can straightforwardly be associated with surveillance pathways, such as, e.g., nonsense-mediated mRNA decay. The main function of these pathways is to reduce errors in gene expression by eliminating mRNA transcripts that contain deletions or premature stop codons (resulting from, for instance, mis-splicing events). However, the easy correlation between decreased mRNA stability and decreased protein stability is not obvious. Pierce and co-workers have identified in HARS2 a mutation in a compound heterozygous state (L200V), which creates an alternative splice-site and leads to an in frame deletion of 12 codons in exon 6 (L200_K211). The level of the spliced-mRNA is significantly increased in the affected child compared to the unaffected (but carrier) father. Transient expression of the spliced-mRNA into HEK293T cells results in a poorly detectable mutant protein, suggesting its instability. However, the mechanism involved in stability deficiency for the mutant protein remains unclear [90]. In three of the PCH6 patients, compound heterozygous mutations I9V/R504_L529, R245Q/R469H, and W241R/Q12R were found in RARS2. It was shown by western blot experiments on cultured fibroblasts that the expression level of the total proteins was reduced down to approximately 28% of the wild-type content. However, the level of mt-ArgRS-encoding mRNAs, as measured by quantitative PCR, remains low but normal in the patients, excluding transcripts instability induced by the mutations [91]. Three mutations (C152F, Q184K, and D560V) within DARS2 were also shown to have an impact on the expression of mt-AspRS. Western blotting on transiently transfected HEK293T cells with mutated sequences indicates strongly reduced steady state levels of the mutant proteins. Further analyses on cycloheximide (to inhibit production of newly synthesized proteins) treated cells indicate a decreased stability of ~50% of the three mutant proteins as compared to the wild-type protein [102].
4.1.3 Defects in mt-aaRSs Import
Several pathology-related mutations are found within (or close to) the predicted MTS. Mutations M1X and R7X, in EARS2, are nonsense and likely lead to untranslated and truncated products, respectively. None will have the opportunity to be imported into mitochondria [89]. Two mutations, Q12R(Q12fsX25) and I9V, are located within the MTS-encoding sequence of RARS2. The mutation Q12R was predicted to enhance import efficiency of mt-ArgRS. As already discussed, this mutation also and mostly interferes with a splicing-enhancer element and causes the retention of 221 bp from intron 1, leading to a frameshift that truncates the protein after residue 25 (Q12fsX25) and likely prevents its import into the mitochondria [87]. The role of the I9V mutation on import of mt-ArgRS into mitochondria is difficult to anticipate since both amino acids are hydrophobic/aliphatic residues, with very similar physico-chemical properties. Predictions on the probability of the I9V mutant to be imported into mitochondria indeed suggest a modest effect of the mutation [91]. Confocal microscopy imaging revealed that mutation S45G located in the predicted MTS of mt-AspRS affects neither the targeting nor the binding of the protein to the mitochondria. However, by combining in vitro import and processing assays, the translocation step was found to be impaired by the mutation [103]. A more recent study investigating the impact of nine mutations found in DARS2 on sub-cellular localization of mt-AspRS by immuno-cytochemistry (using antibodies against transiently expressed tagged mutant proteins in HEK293T cells) did not confirm a localization defect [102].
4.2 Impact of Mutations on mt-aaRSs Function
4.2.1 Impact on mt-aaRSs Enzyme Activities
The housekeeping activity of the aaRSs is to provide aminoacylated-tRNAs (aa-tRNAs) for translation. The effectiveness of aminoacylation can be detected either by in vitro or by in vivo methods (Table 2). ATP-PPi exchange assays and in vitro aminoacylation reactions can be used to establish kinetic parameters k cat, K M for ATP, amino acid, and/or tRNA. As examples, mutations L200V and V368L (HARS2) and I329T (FARS2) were shown to affect ATP-PPi exchange ability of mt-HisRS [90] and mt-PheRS [104], respectively. As an alternative procedure of the ATP-PPi exchange experiment, Cassandrini and co-workers applied a colorimetric-based measurement of Pi production [105]. The authors demonstrated that crude mitochondrial extracts, extracted from cultured skin fibroblasts of patients harboring either R245Q/R469H or W241R/Q12R mutations on mt-ArgRS, have residual activities in Pi formation of only 33% and 19%, respectively. However, they also demonstrate that the level of the protein itself is affected by the mutations [91]. Impacts of mutations on in vitro aminoacylation efficiency of recombinant mt-aaRSs have been investigated at several instances. It has been reported, for example, that mutations within mt-AspRS impact the enzymatic activity (measured as nmol of incorporated amino acid per milligram of enzyme per minute) in a maximum range of ~135-fold (for R263Q, [102]). Other studies have revealed impacts of the Y144C and the F52L mutations on respectively mt-PheRS and mt-TyrRS catalytic efficiencies (relative ratios of k cat/K M, expressed in s−1 μM−1) of 2.3-fold [104] and 9-fold [97]. It should be noted that these effects remain in a moderate range as compared to what has been observed for pathology-related mutations on mt-tRNAs (e.g., >5,000-fold for variants of human mt-tRNALys, [106]).
Measurement of in vivo steady state levels of aa-tRNAs is performed by separation of aminoacylated- and non-aminoacylated-tRNAs on acidic gels. In vivo steady state levels of aa-tRNAs were investigated using total RNA extracted from patients biopsies or cultured cells and northern blotting on acidic gels (reviewed in, e.g., [107]). The total amount of tRNAArg is reduced in patient cells by comparison to the amount found in cells from healthy individuals. They observed, however, that remaining tRNAArg were fully aminoacylated, probably by the few wild-type mt-ArgRS that escaped from the splicing defect engendered by the L13RfsX15 mutation. It is thus suggested that uncharged tRNA might undergo degradation [91, 101]. Belostostky and coworkers showed that the amount of tRNASer(AGY) in cells harboring the D390G mutation in mt-SerRS was reduced to 10–20% as compared to unaffected control, and that the residual pool of tRNASer(AGY) was not aminoacylation. Interestingly, the same mutation affected neither stability nor aminoacylation properties of tRNASer(UCN) [108]. In contrast to these observations, no effect on the steady state level of Met-tRNAMet was observed in patient cells, despite a clear reduced amount of mt-MetRS [98].
Additional methods were developed to investigate the possible impact of mutations on enzyme activity in vivo. For instance, human mutations were modeled on yeast strains, deleted from either the MSR1 or the HTS1 gene (homologues of human RARS2 and HARS2, respectively). Homologue mutants of R469H and R245Q were able to complement MSR1-deleted strains under fermentable conditions (a situation where the respiratory chain is dispensable), but unable (R469H) or barely able (R245Q) to complement MSR1-deleted strain under respiratory conditions. In contrast, a homologue mutant of W241R fully complements the same yeast strain [91]. The homologue mutant L200_K211del was unable to complement a yeast HTS1-deleted strain, and the corresponding human sequence couldn’t be expressed in bacteria or in human cells, suggesting that this mutant is likely to be unable to provide any activity in vivo and confirming the instability of the protein [90]. As an alternate experimental procedure, the retroviral expression of YARS2 rescues the translation defect observed in patient muscle cells [109]. Finally, the correlation between mutations in MARS2 and the ARSAL pathology was confirmed by using the fly as a model organism [98].
As an outcome, defects in the aminoacylation properties of the mt-aaRSs are not necessarily sufficient to explain the pathogenicity of a mutation. Therefore, the cellular environment and additional physiological conditions have to be considered to understand clearly their pathogenic impacts. As examples, tissue-related concentration of the different substrates (e.g., [104, 110]) and/or alternated yet unidentified functions of the mt-aaRSs (e.g., [82, 86, 109]) have been conjectured.
4.2.2 Structure–Function Connections
AaRSs are modular enzymes, composed of well-defined and organized domains, with conserved amino acid residues having either a structural or architectural role, or a function in the chemistry of substrate recognition or in the aminoacylation reaction. Evidence suggests that replacement of key conserved residues may alter essential physico-chemical properties (side chain length, net charge, polarity, hydrophobicity, hydrophilicity, …) and thus may have a key impact on the properties of the protein. Present-day knowledge of the 3D structures of mt-aaRSs, or of aaRSs from evolutionary related species, is of great help for connecting the structural impact of pathology-related mutations with its possible functional consequences. For instance, investigation of the crystallographic structures of prokaryotic HisRSs revealed that the mutation L200 and V368 are both implicated in packing interactions with highly conserved hydrophobic amino acids, involved respectively in ATP binding and in histidine recognition [90]. The two mutations (L200V and V368L) may destabilize the packing interactions, engendering movements that are perturbing some secondary structure elements and possibly reducing binding affinities for either ATP or histidine. In agreement, activities of both mutant proteins, measured by ATP-PPi exchange assay, are significantly reduced relative to the wild-type protein. In a second example, the structural and functional impacts of three pathology-related mutations affecting mt-PheRS could be connected [104]. The crystallographic structure of the enzyme [72, 77] reveals that I329 is located within the ATP-binding site. Replacement of this residue by the small and uncharged threonine should result in a widened ATP-binding site likely decreasing the affinity for the small substrate. In agreement, ATP-PPi exchange kinetic assay confirms a 2.5-fold decreased affinity for ATP for the mutant enzyme, while the binding for phenylalanine remains unaffected. In addition, D391 and Y144 are situated on both sides of the contact surface between the catalytic core and the anticodon-binding domain of mt-PheRS and are stabilizing (by forming hydrogen bounds with key conserved residues) the –closed– state of mt-PheRS. The enzyme was shown to undergo drastic conformational changes upon tRNA binding towards the functional –open– state [72] (Fig. 5). Mutations D391V and Y144C are likely to alter the rotation mechanisms upon tRNA binding and thus to affect the conformational stability of the protein. Along this line, a clear decrease for tRNAPhe binding (increased KM) was observed, but only for the Y144C mutant enzyme. Instead, a decreased affinity for phenylalanine was measured for the D391V mutant, despite D391 not being situated in the catalytic core. As an explanation, the authors are emphasizing that D391 is involved into a close network of interactions encompassing conserved residues of motif 2 (Y188) and near motif 3 (R330). The D391V replacement may cause R330 and other neighboring residues to adopt different conformations, leading to perturbations in a loop, which is critical for binding and coordination of phenylalanine [104]. The ninefold loss of catalytic efficiency (k cat/K M) measured for the F52L mutant of mt-TyrRS by in vitro aminoacylation assay [97] might also be explained by the localization of this residue near the catalytic center, as observed within the crystallographic structure of the enzyme [76].
In contrast, 3D representations of aaRSs are not always sufficient to explain or predict the molecular effect of pathology-related mutations. As previously underlined, the V368L mutation was shown to reduce the enzymatic activity of mt-HisRS, in agreement with its location inside the conserved HisB motif (which is specific to HisRSs and contributes to histidine binding pocket). However, this same mutation was also shown to influence the rate of protein dimerization, although it is not localized at the dimerization interface [90]. Similarly, R592 and L155 are respectively situated in 21.15 Å of a site that could have an editing activity, and in the surrounding of conserved catalytic residues of mt-AlaRS. However, their mutations neither affect editing activity nor aminoacylation properties of the enzyme, leaving the connection between structural predictions and functional mechanisms unclear [110]. Mt-SerRS has the functional peculiarity of being able to recognize two isoacceptor tRNAs (tRNASer AGY and tRNASer UCN). Investigation of the crystallographic structure of bovine mt-SerRS revealed key residues responsible for recognition but also discrimination of the two isoacceptors. Those residues are situated within the helical arm of the synthetase and within or flanking the “distal helix” (Fig. 5), shown to be a structural peculiarity of the mitochondrial enzyme [75]. Analysis of aminoacylation properties revealed that the D390G mutation significantly impacts the acylation of tRNASer AGY but does not alter that of tRNASer UCN. Unexpectedly, the D390 residue is not situated in the isoacceptors discriminating area but in a beta-strand from the catalytic core, far away from the “distal helix” and the helical arm [108].
Despite these few examples where the connection between the structure and the function is not obvious, any future knowledge on crystallographic structures and/or on biophysical properties of mt-aaRSs will be of help to understand the mechanistic aspect and functional impacts of some of the pathology-related mutations. It will also be of help to predict and direct functional investigations. As an example, the crystallographic structure of the yeast cytosolic ArgRS was investigated to assess the possible consequences of the M404K and K158del mutations and to predict a tRNA binding deficiency for the first mutation, and an altered aminoacylation property for the second [92]. Similar predictions could then be drawn for other pathology-related mutations.
4.3 Impact of Mutations on Mitochondrial Translation and Activity of the Respiratory Chain Complexes
As stated above, ATP synthesis is dependent on the coordinated expression of nuclear and mitochondrial genes and requires precise recognition between tRNAs and mt-aaRSs to allow for accurate synthesis of aa-tRNAs. Accordingly, key links between the aminoacylation activity of mt-aaRSs in charge of the synthesis of the 13 mt-DNA-encoded proteins and the activity of respiratory chain (RC) complexes are foreseen. It can be anticipated that any dysfunction of a single macromolecule of the translation machinery may have severe impacts either on the translation and/or on the activity of the mt-DNA-encoded RC subunits (for all complexes except Complex II, of complete nuclear origin). However, this view appears too simplistic. Figure 8 summarizes observed molecular defects (at the levels of mRNAs expression and mt-aaRSs biosynthesis and functioning) and possible defects in the activity of the RC complexes for all reported mutations. It also schematizes that the routes linking the molecular impact of a mutation with its possible phenotypic effect are not yet always fully deciphered.
Summary of the impacts of pathology-related mutations on the different steps of mt-aaRS life cycle and subsequent products activities. Mt-aaRSs (on the outer circle) are represented using the one-letter code. Mt-aaRSs affected by pathology-related mutations are in green; those yet unrelated to human pathologies are in gray. Observed molecular defects at the levels of mRNAs expression and mt-aaRSs biosynthesis and functioning are represented in blue in the middle circle. Possible defects on activity of the RC complexes are schematized in orange in the inner circle. The routes that link the molecular impact of a mutation with its possible phenotypic effect and lead to mitochondrial disorders are schematized (plain and dashed arrows). Mutations displayed in red were shown to engender obvious connections between mt-aaRS expression and/or aminoacylation defects with RC dysfunctioning. However, these routes are diverse, and interruptions indicate either the absence of full investigation, or the absence of clear molecular link (discontinuous or dashed arrows)
4.3.1 Translation and/or Activity of the Respiratory Chain Complexes Sub-units are Differentially Affected
Defects in the translation of mt-DNA-encoded RC subunits were reported to be correlated with mutations within YARS2 [97, 111], MARS2 [98], and FARS2 [104]. In addition, defects in the activity of those complexes were reported to be correlated with mutations within YARS2 [97, 111], SARS2 [108], RARS2 [92, 101], MARS2 [98], FARS2 [104], EARS2 [89], DARS2 [112], and AARS2 [110]. However, affected subunits may vary from one case to another. Combinations of translation and/or activity defects are numerous. For example, the translation of all complexes is affected by mutation in YARS2, while solely the expression of Complex IV is impaired by mutations in FARS2. Similarly, the activity of all the complexes was shown to be impaired by a mutation in SARS2, while only Complex IV is moderately affected by the M404K/K158del mutations in RARS2, leaving unaffected the activities of Complexes I and III. To be noticed as well, gene rearrangement of MARS2 engenders defects in the translation of all subunits, but noticeably causes activity defects solely in Complex I.
4.3.2 The Same Affected Gene, but Different Impacts on Translation and/or Activity of the Sub-units
A striking observation is that mutations affecting the same gene have different consequences. For instance, mutations M342V/Q12R and IVS2 + 5(A>G)/K291R/K568K, both located in RARS2, respectively do not affect and drastically impair respiratory chain complexes [92, 101]. More confusingly, the same combination of mutations (IVS2 + 5(A>G)/K291R/K568K) does not engender the same effects on three affected individuals. This combination of mutations reduced activities of Complexes I, III, and IV in muscles from patient II-2, but reduced activity of Complexes I in muscles from patient II-4, or activity of Complex IV from patient II-5 [87].
4.3.3 Tissues Specificity
Phenotypic manifestations of mutations have been investigated several times on mitochondria extracted from cell cultures established from biopsies specimens obtained from patients. Here again, discrepancies are observed depending on the investigated tissues. Fibroblasts are frequently used, although investigations performed on those cells showed effects on RC subunits solely for mutations within EARS2 [89]. Conversely, measurements made on muscle, brain, or heart cells show defects on RC subunits at several instances (muscle cells affected by DARS2, RARS2, YARS2, SARS2, AARS2, EARS2, and FARS2 mutations, brain cells affected by AARS2 and FARS2 mutations, and heart cells affected by AARS2 mutations). Consequently, discrepancies can be observed when testing the effect of a given mutation on different types of cells. For instance, the F52L mutation found in YARS2 does not show any impact on mitochondrial translation in fibroblasts, but engenders drastic impairment in skeletal muscle and myotubes [97]. Similarly, mutations in AARS2 affect the RC activity in only muscle and brain cells, but not in liver cells [110]. Of note, it is not yet possible to make a direct link with the tissue prevalence of the disease (see Table 3) since cellular models haven’t been systematically investigated.
In summary, the diverse effects of mutations on the respiratory chain complexes highlight the numerous routes linking the molecular impact of a mutation with its possible phenotypic effect. These routes are summarized and schematized in Fig. 8. Some examples show obvious connections between mt-aaRS expression and/or aminoacylation defects with RC dysfunctioning (e.g., mutations in MARS2, YARS2, and SARS2). Conversely, it can be seen that, for some of the reported cases, investigations are not yet complete so that it is not possible to draw the pathogenic pathway (e.g., mutations in HARS2, EARS2, AARS2, DARS2, RARS2, and FARS2). Another situation is visible in the scheme, where the molecular routes are diverse for the set of mutations affecting the same gene (mutations in, e.g., DARS2, RARS2, and FARS2). It can thus be hypothesized that a diversity of mechanisms, with tissue specificity, contributes to the heterogeneous manifestations of mitochondrial disorders.
5 Outlook
Human mitochondrial aminoacylation systems deserve specific attention as a consequence of their recently recognized connection with human pathologies. Understanding the fundamental properties/peculiarities of aminoacylation systems is critical to resolving the link between mutation and pathology. In the past 20 years a large number of human neuromuscular and neurodegenerative disorders have been reported to be correlated with point mutations in the mt-mRNAs, mt-rRNAs, and especially in mt-tRNAs (reviewed in, e.g., [5, 6, 10, 113–115]). Among the mutations leading to “mitochondrial disorders,” more than 230 are distributed all over the 22 tRNAs. Numerous studies have attempted to unravel the molecular impacts of the mutations on the various properties of the tRNA that lead to a mosaïcity of phenotypic effects. Whilst there is no general rule, a trend towards a structural perturbation as initial molecular impact of mutations can be retained [116, 117]. However, the housekeeping function of tRNAs, namely their capacity to become esterified by an amino acid, is not systematically affected in mutated variants, so that alternative functions of tRNAs or at least alternative partnerships have to be considered [118]. The discovery of a new family of impacted genes, the mt-aaRSs, further extends the complexity of mitochondrial disorders.
The distance between the birth of an mt-aaRS and its final role in ATP synthesis is very long, so that the gap between the comprehension of the molecular impact of mutations in macromolecules and a dysfunction of the respiratory chain is very large. Also, considering the sole/common outcome from any defect in mt-aaRSs (or in mt-tRNAs) results from an effect on the translation rate of each of the 13 mt-DNA-encoded proteins is a view that rapidly appeared too simplistic. Indeed, the major outcome from the present review is that there is no common combination of affected steps that correlates the 64 reported cases (combining 65 mutations within nuclear-encoded mt-aaRS genes) to the various observed phenotypic expressions. There is obviously no “favored” mt-aaRS gene. Nine are reported today, but their recent, rapid and exponential correlations with human pathologies suggest as evidence that all mt-aaRS genes are likely to be affected by pathology-related mutations, but remain to be revealed. There are also no “favored” affected parts of the protein, which is in agreement with the fact that all steps of the mt-aaRS life cycle can be impacted. Finally, despite the fact that primary observations would suggest an exclusive connection of mt-aaRS disorders to the nervous system and to inherited neurological diseases, sporadic manifestations were lately observed in, e.g., skeletal muscle, kidney, lung, and/or heart. Thus, links between the activity of a given aaRS to mitochondrial translation on one hand and ATP production on the other, involve a number of issues that need to be further explored. Those issues may consider a possible combined effect of mutations affecting other gene(s), such as, for instance, affecting the tRNA modifying enzyme as observed in MLASA patients [109]. Those issues should also take into account the possibility that aminoacylation may turn out not to be the sole function of mt-aaRSs in a living cell, and that these enzymes may also participate in other processes and/or be implicated in various fine-tuning mechanisms, as already shown for various bacterial and eukaryal aaRSs; see below. Thus it becomes obvious that we have to integrate mt-aaRSs into a functional network at the cellular level. In other words, it is of outstanding interest to nail down all the potential interacting components of mt-aaRSs and study their dynamic location within the cell.
In support of this assumption, developments in genomics and post-genomics, associated with conventional biochemical studies, led to the finding of unexpected non-conventional auxiliary functions for human cytosolic aaRSs and connections to other cellular activities (reviewed in, e.g., [119]). Examples include enzymes secreted as procytokines that, after activation, operate in pathways linked to the immune system or angiogenesis (e.g., cyt-TyrRS, cyt-TrpRS [120, 121]), or involved in the vascular development (cyt-SerRS [122]). In addition, accumulating evidence indicates that disruption of non-canonical functions of cytosolic aaRSs connects to various types of diseases, including neural pathologic conditions and cancer [123]. For example, point mutations in human cytosolic TyrRS and cytosolic GlyRS are associated with Charcot-Marie-Tooth (CMT) diseases. Examination of the aminoacylation activities demonstrates that CMT disease can occur without loss of aminoacylation activity [124, 125]. Finally, nowadays evidence indicates that macromolecular assemblies might be sources for proteins with auxiliary functions. Multiprotein complexes containing aaRSs are widely found in all three domains of life, playing roles in apoptosis, viral assembly, and regulation of transcription and translation (reviewed in [126]. As an example, the cytosolic translational apparatus in human cells is highly organized. Nine of the cytosolic aaRS are assembled into the MARS complex (with three auxiliary proteins), which has been emphasized as an anchoring platform for multitasking proteins [127–129]. Those were shown to be recognizable not only by displaying atypical functional activities (possibly linked to structural inventions [69, 119, 130]), but also by their atypical cellular organization, atypical selection pressure, or by atypical “omics” behaviors. The organization of the aaRS within human mitochondria remains mostly unknown at present but will merit full attention in the near future.
Abbreviations
- AaRS:
-
Aminoacyl-tRNA synthetase (specificity is indicated by the name of the amino acid (abbreviated in a three-letter code) transferred to the cognate tRNA. As an example, AspRS stands for aspartyl-tRNA synthetase)
- mt:
-
Mitochondrial
- MTS:
-
Mitochondrial targeting sequence
- RC:
-
Respiratory chain
References
Scheffler IE (2001) A century of mitochondrial research: achievements and perspectives. Mitochondrion 1:3–31
Anderson S, Bankier AT, Barrell BG et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290:457–465
Ojala D, Montoya J, Attardi G (1981) tRNA punctuation model of RNA processing in human mitochondria. Nature 290:470–474
Christian BE, Spremulli LL (2012) Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 1819:1035–1054
Florentz C, Sohm B, Tryoen-Tóth P et al (2003) Human mitochondrial tRNAs in health and disease. Cell Mol Life Sci 60:1356–1375
Suzuki T, Nagao A, Suzuki T (2011) Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 45:299–329
Watanabe K (2010) Unique features of animal mitochondrial translation systems. The non-universal genetic code, unusual features of the translational apparatus and their relevance to human mitochondrial diseases. Proc Jpn Acad Ser B Phys Biol Sci 86:11–39
Bonnefond L, Fender A, Rudinger-Thirion J et al (2005) Toward the full set of human mitochondrial aminoacyl-tRNA synthetases: characterization of AspRS and TyrRS. Biochemistry 44:4805–4816
Sissler M, Pütz J, Fasiolo F, Florentz C (2005) Mitochondrial aminoacyl-tRNA synthetases. In: Ibba M, Francklyn C, Cusack S (eds) Aminoacyl-tRNA synthetases. Landes Biosciences, Georgetown, pp 271–284
Ylikallio E, Suomalainen A (2012) Mechanisms of mitochondrial diseases. Ann Med 44:41–59
Shoffner JM, Lott MT, Lezza AM et al (1990) Myoclonic epilepsy and ragged-red fiber disease (MERRF) is associated with a mitochondrial DNA tRNA(Lys) mutation. Cell 61:931–937
Goto Y, Nonaka I, Horai S (1990) A mutation in the tRNALeu(UUR) gene associated with the MELAS subgroup of mitochondrial encephalomyopathies. Nature 348:651–653
Dimauro S, Davidzon G (2005) Mitochondrial DNA and disease. Ann Med 37:222–232
Diaz F (2010) Cytochrome c oxidase deficiency: patients and animal models. Biochim Biophys Acta 1802:100–110
Stumpf JD, Copeland WC (2011) Mitochondrial DNA replication and disease: insights from DNA polymerase γ mutations. Cell Mol Life Sci 68:219–233
Scheper GD, Van der Knaap MS, Proud CG (2007) Translation matters: protein synthesis defects in inherited disease. Nat Rev Genet 8:711–723
Konovalova S, Tyynismaa H (2013) Mitochondrial aminoacyl-tRNA synthetases in human disease. Mol Genet Metab 108:206–211
Reeve AK, Krishnan KJ, Turnbull D (2009) Mitochondrial DNA mutations in disease, aging, and neurodegeneration. Ann N Y Acad Sci 1147:21–29
Wallace DC (2010) Mitochondrial DNA mutations in disease and aging. Environ Mol Mutagen 51:440–450
McFarland R, Elson JL, Taylor RW et al (2004) Assigning pathogenicity to mitochondrial tRNA mutations: when “definitely maybe” is not good enough. Trends Genet 20:591–596
Zeviani M, Di Donato S (2004) Mitochondrial disorders. Brain 127:2153–2172
Smits P, Smeitink J, Van den Heuvel L (2010) Mitochondrial translation and beyond: processes implicated in combined oxidative phosphorylation deficiencies. J Biomed Biotechnol 2010:737385
Fernández-Silva P, Acín-Pérez R, Fernández-Vizarra E et al (2007) In vivo and in organello analyses of mitochondrial translation. Methods Cell Biol 80:571–588
DiMauro S, Hirano M (2003) Mitochondrial DNA deletion syndromes. In: Pagon RA, Bird TD, Dolan CR, Stephens K, Adm MP (eds) GeneReviews, Seattle
Kunz WS, Kudin A, Vielhaber S et al (2000) Flux control of cytochrome c oxidase in human skeletal muscle. J Biol Chem 275:27741–27745
Munnich A, Rustin P (2001) Clinical spectrum and diagnosis of mitochondrial disorders. Am J Med Genet 106:4–17
Barrientos A (2002) In vivo and in organello assessment of OXPHOS activities. Methods 26:307–316
Brand MD, Nicholls DG (2011) Assessing mitochondrial dysfunction in cells. Biochem J 435:297–312
Chance B, Williams GR (1955) A simple and rapid assay of oxidative phosphorylation. Nature 175:1120–1121
N’Guessan B, Zoll J, Ribera F et al (2004) Evaluation of quantitative and qualitative aspects of mitochondrial function in human skeletal and cardiac muscles. Mol Cell Biochem 256–257:267–280
Veksler VI, Kuznetsov AV, Sharov VG et al (1987) Mitochondrial respiratory parameters in cardiac tissue: a novel method of assessment by using saponin-skinned fibers. Biochim Biophys Acta 892:191–196
Letellier T, Malgat M, Coquet M et al (1992) Mitochondrial myopathy studies on permeabilized muscle fibers. Pediatr Res 32:17–22
Bouitbir J, Charles A-L, Echaniz-Laguna A et al (2012) Opposite effects of statins on mitochondria of cardiac and skeletal muscles: a “mitohormesis” mechanism involving reactive oxygen species and PGC-1. Eur Heart J 33:1397–1407
Nijtmans LG, Henderson NS, Holt IJ (2002) Blue native electrophoresis to study mitochondrial and other protein complexes. Methods 26:327–334
Brown WM, George M Jr, Wilson AC (1979) Rapid evolution of animal mitochondrial DNA. Proc Natl Acad Sci USA 76:1967–1971
Castellana S, Vicario S, Saccone C (2011) Evolutionary patterns of the mitochondrial genome in Metazoa: exploring the role of mutation and selection in mitochondrial protein coding genes. Genome Biol Evol 3:1067–1079
Giegé R, Jühling F, Pütz J et al (2012) Structure of transfer RNAs: similarity and variability. Wiley Interdiscip Rev RNA 3:37–61
Helm M, Brulé H, Friede D et al (2000) Search for characteristic structural features of mammalian mitochondrial tRNAs. RNA 6:1356–1379
Mudge SJ, Williams JH, Eyre HJ et al (1998) Complex organisation of the 5′-end of the human glycine tRNA synthetase gene. Gene 209:45–50
Shiba K, Schimmel P, Motegi H, Noda T (1994) Human glycyl-tRNA synthetase. Wide divergence of primary structure from bacterial counterpart and species-specific aminoacylation. J Biol Chem 269:30049–30055
Tolkunova E, Park H, Xia J et al (2000) The human lysyl-tRNA synthetase gene encodes both the cytoplasmic and mitochondrial enzymes by means of an unusual alternative splicing of the primary transcript. J Biol Chem 275:35063–35069
Rinehart J, Krett B, Rubio MA et al (2005) Saccharomyces cerevisiae imports the cytosolic pathway for Gln-tRNA synthesis into the mitochondrion. Genes Dev 19:583–592
Ibba M, Soll D (2000) Aminoacyl-tRNA synthesis. Annu Rev Biochem 69:617–650
Pujol C, Bailly M, Kern D et al (2008) Dual-targeted tRNA-dependent amidotransferase ensures both mitochondrial and chloroplastic Gln-tRNAGln synthesis in plants. Proc Natl Acad Sci USA 105:6481–6485
Schön A, Kannangara CG, Gough S, Söll D (1988) Protein biosynthesis in organelles requires misaminoacylation of tRNA. Nature 331:187–190
Frechin M, Duchêne A-M, Becker HD (2009) Translating organellar glutamine codons: a case by case scenario? RNA Biol 6:31–34
Nagao A, Suzuki T, Katoh T et al (2009) Biogenesis of glutaminyl-mt tRNAGln in human mitochondria. Proc Natl Acad Sci USA 106:16209–16214
Frechin M, Senger B, Brayé M et al (2009) Yeast mitochondrial Gln-tRNA(Gln) is generated by a GatFAB-mediated transamidation pathway involving Arc1p-controlled subcellular sorting of cytosolic GluRS. Genes Dev 23:1119–1130
Alfonzo JD, Söll D (2009) Mitochondrial tRNA import–the challenge to understand has just begun. Biol Chem 390:717–722
Gray MW, Burger G, Lang BF (1999) Mitochondrial evolution. Science 283:1476–1481
Brindefalk B, Viklund J, Larsson D et al (2007) Origin and evolution of the mitochondrial aminoacyl-tRNA synthetases. Mol Biol Evol 24:743–756
Pfanner N (2000) Protein sorting: recognizing mitochondrial presequences. Curr Biol 10:R412–R415
Baker MJ, Frazier AE, Gulbis JM, Ryan MT (2007) Mitochondrial protein-import machinery: correlating structure with function. Trends Cell Biol 17:456–464
Becker T, Böttinger L, Pfanner N (2012) Mitochondrial protein import: from transport pathways to an integrated network. Trends Biochem Sci 37:85–91
Bolender N, Sickmann A, Wagner R et al (2008) Multiple pathways for sorting mitochondrial precursor proteins. EMBO Rep 9:42–49
Gakh O, Cavadini P, Isaya G (2002) Mitochondrial processing peptidases. Biochim Biophys Acta 1592:63–77
Van der Laan M, Hutu DP, Rehling P (2010) On the mechanism of preprotein import by the mitochondrial presequence translocase. Biochim Biophys Acta 1803:732–739
Neupert W, Herrmann JM (2007) Translocation of proteins into mitochondria. Annu Rev Biochem 76:723–749
Schmidt O, Pfanner N, Meisinger C (2010) Mitochondrial protein import: from proteomics to functional mechanisms. Nat Rev Mol Cell Biol 11:655–667
Vögtle F-N, Wortelkamp S, Zahedi RP et al (2009) Global analysis of the mitochondrial N-proteome identifies a processing peptidase critical for protein stability. Cell 139:428–439
Varshavsky A (2011) The N-end rule pathway and regulation by proteolysis. Protein Sci 20(8):1298–1345
Sissler M, Lorber B, Messmer M et al (2008) Handling mammalian mitochondrial tRNAs and aminoacyl-tRNA synthetases for functional and structural characterization. Methods 44:176–189
Bullard JM, Cai YC, Spremulli LL (2000) Expression and characterization of the human mitochondrial leucyl-tRNA synthetase. Biochim Biophys Acta 1490:245–258
Yao Y-N, Wang L, Wu X-F, Wang E-D (2003) Human mitochondrial leucyl-tRNA synthetase with high activity produced from Escherichia coli. Protein Expr Purif 30:112–116
Gaudry A, Lorber B, Neuenfeldt A et al (2012) Re-designed N-terminus enhances expression, solubility and crystallizability of mitochondrial protein. Protein Eng Des Sel 25:473–481
Neuenfeldt A, Lorber B, Ennifar E et al (2012) Thermodynamic properties distinguish human mitochondrial aspartyl-tRNA synthetase from bacterial homolog with same 3D architecture. Nucleic Acids Res 41:2698–2708
Cusack S, Berthet-Colominas C, Härtlein M et al (1990) A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5Å. Nature 347:249–255
Eriani G, Delarue M, Poch O et al (1990) Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature 347:203–206
Guo M, Schimmel P, Yang X-L (2010) Functional expansion of human tRNA synthetases achieved by structural inventions. FEBS Lett 584:434–442
Bullard JM, Cai YC, Demeler B, Spremulli LL (1999) Expression and characterization of a human mitochondrial phenylalanyl-tRNA synthetase. J Mol Biol 288:567–577
Sanni A, Walter P, Boulanger Y et al (1991) Evolution of aminoacyl-tRNA synthetase quaternary structure and activity: Saccharomyces cerevisiae mitochondrial phenylalanyl-tRNA synthetase. Proc Natl Acad Sci USA 88:8387–8391
Klipcan L, Moor N, Finarov I et al (2012) Crystal structure of human mitochondrial PheRS complexed with tRNAPhe in the active “open” state. J Mol Biol 415:527–537
Kaiser E, Hu B, Becher S et al (1994) The human EPRS locus (formerly the QARS locus): a gene encoding a class I and a class II aminoacyl-tRNA synthetase. Genomics 19:280–290
Bhat TN, Blow DM, Brick P, Nyborg J (1982) Tyrosyl-tRNA synthetase forms a mononucleotide-binding fold. J Mol Biol 158:699–709
Chimnaronk S, Gravers Jeppesen M, Suzuki T et al (2005) Dual-mode recognition of noncanonical tRNAsSer by seryl-tRNA synthetase in mammalian mitochondria. EMBO J 24:3369–3379
Bonnefond L, Frugier M, Touzé E et al (2007) Crystal structure of human mitochondrial tyrosyl-tRNA synthetase reveals common and idiosyncratic features. Structure 15:1505–1516
Klipcan L, Levin I, Kessler N et al (2008) The tRNA-induced conformational activation of human mitochondrial phenylalanyl-tRNA synthetase. Structure 16:1095–1104
Fender A, Sauter C, Messmer M et al (2006) Loss of a primordial identity element for a mammalian mitochondrial aminoacylation system. J Biol Chem 281:15980–15986
Messmer M, Blais SP, Balg C et al (2009) Peculiar inhibition of human mitochondrial aspartyl-tRNA synthetase by adenylate analogs. Biochimie 91:596–603
Fender A, Gaudry A, Jühling F et al (2012) Adaptation of aminoacylation identity rules to mammalian mitochondria. Biochimie 94:1090–1097
Kumazawa Y, Himeno H, Miura K, Watanabe K (1991) Unilateral aminoacylation specificity between bovine mitochondria and eubacteria. J Biochem 109:421–427
Scheper GC, Van der Klok T, Van Andel RJ et al (2007) Mitochondrial aspartyl-tRNA synthetase deficiency causes leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation. Nat Genet 39:534–539
Isohanni P, Linnankivi T, Buzkova J et al (2010) DARS2 mutations in mitochondrial leucoencephalopathy and multiple sclerosis. J Med Genet 47:66–70
Labauge P, Dorboz I, Eymard-Pierre E et al (2011) Clinically asymptomatic adult patient with extensive LBSL MRI pattern and DARS2 mutations. J Neurol 258:335–337
Lin J, Chiconelli Faria E, Da Rocha AJ et al (2010) Leukoencephalopathy with brainstem and spinal cord involvement and normal lactate: a new mutation in the DARS2 gene. J Child Neurol 25:1425–1428
Namavar Y, Barth PG, Kasher PR et al (2011) Clinical, neuroradiological and genetic findings in pontocerebellar hypoplasia. Brain 134:143–156
Rankin J, Brown R, Dobyns WB et al (2010) Pontocerebellar hypoplasia type 6: a British case with PEHO-like features. Am J Med Genet A 152A:2079–2084
Sharma S, Sankhyan N, Kumar A et al (2011) Leukoencephalopathy with brain stem and spinal cord involvement and high lactate: a genetically proven case without elevated white matter lactate. J Child Neurol 26:773–776
Steenweg ME, Ghezzi D, Haack T et al (2012) Leukoencephalopathy with thalamus and brainstem involvement and high lactate ‘LTBL' caused by EARS2 mutations. Brain 135:1387–1394
Pierce SB, Chisholm KM, Lynch ED et al (2011) Mutations in mitochondrial histidyl tRNA synthetase HARS2 cause ovarian dysgenesis and sensorineural hearing loss of Perrault syndrome. Proc Natl Acad Sci USA 108:6543–6548
Cassandrini D, Cilio MR, Bianchi M et al (2012) Pontocerebellar hypoplasia type 6 caused by mutations in RARS2: definition of the clinical spectrum and molecular findings in five patients. J Inherit Metab Dis 36:43–53
Glamuzina E, Brown R, Hogarth K et al (2012) Further delineation of pontocerebellar hypoplasia type 6 due to mutations in the gene encoding mitochondrial arginyl-tRNA synthetase, RARS2. J Inherit Metab Dis 35:459–467
Antonellis A, Green ED (2008) The role of aminoacyl-tRNA synthetases in genetic diseases. Annu Rev Genomics Hum Genet 9:87–107
Miyake N, Yamashita S, Kurosawa K et al (2011) A novel homozygous mutation of DARS2 may cause a severe LBSL variant. Clin Genet 80:293–296
Synofzik M, Schicks J, Lindig T et al (2011) Acetazolamide-responsive exercise-induced episodic ataxia associated with a novel homozygous DARS2 mutation. J Med Genet 48:713–715
Van Berge L, Dooves S, Van Berkel CG et al (2012) Leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation is associated with cell-type-dependent splicing of mtAspRS mRNA. Biochem J 441:955–962
Riley LG, Cooper S, Hickey P et al (2010) Mutation of the mitochondrial tyrosyl-tRNA synthetase gene, YARS2, causes myopathy, lactic acidosis, and sideroblastic anemia–MLASA syndrome. Am J Hum Genet 87:52–59
Bayat V, Thiffault I, Jaiswal M et al (2012) Mutations in the mitochondrial methionyl-tRNA synthetase cause a neurodegenerative phenotype in flies and a recessive ataxia (ARSAL) in humans. PLoS Biol 10:e1001288
Mierzewska H, Van der Knaap MS, Scheper GC et al (2011) Leukoencephalopathy with brain stem and spinal cord involvement and lactate elevation in the first Polish patient. Brain Dev 33:713–717
Yamashita S, Miyake N, Matsumoto N et al (2012) Neuropathology of leukoencephalopathy with brainstem and spinal cord involvement and high lactate caused by a homozygous mutation of DARS2. Brain Dev 35:312–316
Edvardson S, Shaag A, Kolesnikova O et al (2007) Deleterious mutation in the mitochondrial arginyl-transfer RNA synthetase gene is associated with pontocerebellar hypoplasia. Am J Hum Genet 81:857–862
Van Berge L, Kevenaar J, Polder E et al (2012) Pathogenic mutations causing LBSL affect mitochondrial aspartyl-tRNA synthetase in diverse ways. Biochem J 450:345–350
Messmer M, Florentz C, Schwenzer H et al (2011) A human pathology-related mutation prevents import of an aminoacyl-tRNA synthetase into mitochondria. Biochem J 433:441–446
Elo JM, Yadavalli SS, Euro L et al (2012) Mitochondrial phenylalanyl-tRNA synthetase mutations underlie fatal infantile Alpers encephalopathy. Hum Mol Genet 21:4521–4529
Chang GG, Pan F, Yeh C, Huang TM (1983) Colorimetric assay for aminoacyl-tRNA synthetases. Anal Biochem 130:171–176
Sissler M, Helm M, Frugier M et al (2004) Aminoacylation properties of pathology-related human mitochondrial tRNA(Lys) variants. RNA 10:841–853
Köhrer C, Rajbhandary UL (2008) The many applications of acid urea polyacrylamide gel electrophoresis to studies of tRNAs and aminoacyl-tRNA synthetases. Methods 44:129–138
Belostotsky R, Ben-Shalom E, Rinat C et al (2011) Mutations in the mitochondrial seryl-tRNA synthetase cause hyperuricemia, pulmonary hypertension, renal failure in infancy and alkalosis, HUPRA syndrome. Am J Hum Genet 88:193–200
Sasarman F, Nishimura T, Thiffault I, Shoubridge EA (2012) A novel mutation in YARS2 causes myopathy with lactic acidosis and sideroblastic anemia. Hum Mutat 33:1201–1206
Götz A, Tyynismaa H, Euro L et al (2011) Exome sequencing identifies mitochondrial alanyl-tRNA synthetase mutations in infantile mitochondrial cardiomyopathy. Am J Hum Genet 88:635–642
Sasarman F, Karpati G, Shoubridge EA (2002) Nuclear genetic control of mitochondrial translation in skeletal muscle revealed in patients with mitochondrial myopathy. Hum Mol Genet 11:1669–1681
Orcesi S, La Piana R, Uggetti C et al (2011) Spinal cord calcification in an early-onset progressive leukoencephalopathy. J Child Neurol 26:876–880
Rötig A (2011) Human diseases with impaired mitochondrial protein synthesis. Biochim Biophys Acta 1807:1198–1205
Wallace DC (1999) Mitochondrial diseases in man and mouse. Science 283:1482–1488
Yarham JW, Elson JL, Blakely EL et al (2010) Mitochondrial tRNA mutations and disease. Wiley Interdiscip Rev RNA 1:304–324
Levinger L, Mörl M, Florentz C (2004) Mitochondrial tRNA 3′ end metabolism and human disease. Nucleic Acids Res 32:5430–5441
Wittenhagen LM, Kelley SO (2003) Impact of disease-related mitochondrial mutations on tRNA structure and function. Trends Biochem Sci 28:605–611
Jacobs HT, Holt IJ (2000) The np 3243 MELAS mutation: damned if you aminoacylate, damned if you don’t. Hum Mol Genet 9:463–465
Guo M, Schimmel P (2013) Essential nontranslational functions of tRNA synthetases. Nat Chem Biol 9:145–153
Wakasugi K, Slike BM, Hood J et al (2002) Induction of angiogenesis by a fragment of human tyrosyl-tRNA synthetase. J Biol Chem 277:20124–20126
Wakasugi K, Slike BM, Hood J et al (2002) A human aminoacyl-tRNA synthetase as a regulator of angiogenesis. Proc Natl Acad Sci USA 99:173–177
Kawahara A, Stainier DYR (2009) Noncanonical activity of seryl-transfer RNA synthetase and vascular development. Trends Cardiovasc Med 19:179–182
Park SG, Schimmel P, Kim S (2008) Aminoacyl tRNA synthetases and their connections to disease. Proc Natl Acad Sci 105:11043–11049
Seburn KL, Nangle LA, Cox GA et al (2006) An active dominant mutation of glycyl-tRNA synthetase causes neuropathy in a Charcot-Marie-Tooth 2D mouse model. Neuron 51:715–726
Storkebaum E, Leitão-Gonçalves R, Godenschwege T et al (2009) Dominant mutations in the tyrosyl-tRNA synthetase gene recapitulate in Drosophila features of human Charcot-Marie-Tooth neuropathy. Proc Natl Acad Sci USA 106:11782–11787
Hausmann CD, Ibba M (2008) Aminoacyl-tRNA synthetase complexes: molecular multitasking revealed. FEMS Microbiol Rev 32:705–721
Han JM, Lee MJ, Park SG et al (2006) Hierarchical network between the components of the multi-tRNA synthetase complex: implications for complex formation. J Biol Chem 281:38663–38667
Kaminska M, Havrylenko S, Decottignies P et al (2009) Dissection of the structural organization of the aminoacyl-tRNA synthetase complex. J Biol Chem 284:6053–6060
Ray PS, Arif A, Fox PL (2007) Macromolecular complexes as depots for releasable regulatory proteins. Trends Biochem Sci 32:158–164
Guo M, Yang X-L, Schimmel P (2010) New functions of aminoacyl-tRNA synthetases beyond translation. Nat Rev Mol Cell Biol 11:668–674
Uluc K, Baskan O, Yildirim KA et al (2008) Leukoencephalopathy with brain stem and spinal cord involvement and high lactate: a genetically proven case with distinct MRI findings. J Neurol Sci 273:118–122
Tzoulis C, Tran GT, Gjerde IO et al (2012) Leukoencephalopathy with brainstem and spinal cord involvement caused by a novel mutation in the DARS2 gene. J Neurol 259:292–296
Talim B, Pyle A, Griffin H et al (2013) Multisystem fatal infantile disease caused by a novel homozygous EARS2 mutation. Brain 136:e228
Shamseldin HE, Alshammari M, Al-Sheddi T et al (2012) Genomic analysis of mitochondrial diseases in a consanguineous population reveals novel candidate disease genes. J Med Genet 49:234–241
Acknowledgements
We thank Redmond Smyth for many stylistic improvements of the manuscript. Our work is supported by Centre National de la Recherche Scientifique (CNRS), Université de Strasbourg (UdS), and the French National Program “Investissement d’Avenir” (Labex MitCross), administered by the “Agence National de la Recherche,” and referenced ANR-10-IDEX-002-02. The ADIRAL association is acknowledged. HS was supported by Région Alsace, Université de Strasbourg, Association Française contre les Mytopathies (AFM) and Fondation des Treilles.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2013 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Schwenzer, H., Zoll, J., Florentz, C., Sissler, M. (2013). Pathogenic Implications of Human Mitochondrial Aminoacyl-tRNA Synthetases. In: Kim, S. (eds) Aminoacyl-tRNA Synthetases in Biology and Medicine. Topics in Current Chemistry, vol 344. Springer, Dordrecht. https://doi.org/10.1007/128_2013_457
Download citation
DOI: https://doi.org/10.1007/128_2013_457
Published:
Publisher Name: Springer, Dordrecht
Print ISBN: 978-94-017-8700-0
Online ISBN: 978-94-017-8701-7
eBook Packages: Chemistry and Materials ScienceChemistry and Material Science (R0)