Abstract
The publication of statistical results based on the use of computational tools requires that the data as well as the code are provided in order to allow to reproduce and verify the results with reasonable effort. However, this only allows to rerun the exact same analysis. While this is helpful to understand and retrace the steps of the analysis which led to the published results, it constitutes only a limited proof of reproducibility. In fact for “true” reproducibility one might require that the essentially same results are obtained in an independent analysis. To check for this “true” reproducibility of results of a text mining application we replicate a study where a latent Dirichlet allocation model was fitted to the document-term matrix derived for the abstracts of the papers published in the Proceedings of the National Academy of Sciences from 1991 to 2001. Comparing the results we assess (1) how well the corpus and the document-term matrix can be reconstructed, (2) if the same model would be selected and (3) if the analysis of the fitted model leads to the same main conclusions and insights. Our study indicates that the results from this study are robust with respect to slightly different preprocessing steps and the use of a different software to fit the model.
Access provided by Autonomous University of Puebla. Download chapter PDF
Similar content being viewed by others
Keywords
1 Introduction
Reproducibility of research results is a topic which has recently received increased interest [6, 8]. To ensure easy reproducibility of statistical analyses, data and code are often made available. This allows to rerun the exact same procedures using in general the complete same software environment in order to arrive at the same results [9]. However, as Keiding points out “it ridicules our profession to believe that there is a serious check on reproducibility in seeing if somebody else’s computer reaches the same conclusion using the same code on the same data set as the original statistician’s computer did [7, p. 377].” True reproducibility therefore would require that an independent analysis arrives at the same results and conclusions, i.e., one might only claim that a result is reproducible, when approximately the same results are obtained if the data preprocessing as well as the model fitting steps are essentially the same, but not necessarily identical. This would imply that the results are robust to small changes in the data preprocessing and model fitting process.
In the following we perform an independent reanalysis of the text mining application published by Griffiths and Steyvers in 2004 [4, in the following referred to as GS2004]. GS2004 use the latent Dirichlet allocation [LDA, 1] model with collapsed Gibbs sampling to analyze the abstracts of papers published in the Proceedings of the National Academy of Sciences (PNAS) from 1991 to 2001. LDA was introduced by Blei and co-authors as a generative probabilistic model for collections of discrete data such as text corpora. Because PNAS is a multidisciplinary, peer-reviewed scientific journal with a high impact factor, this corpus should allow to discover some of the topics addressed by scientific research in this time period.
We try to reproduce the results presented in GS2004 using open-source software with respect to (1) retrieving and preprocessing the corpus to construct the document-term matrix and (2) fitting the LDA model using collapsed Gibbs sampling. In our approach we rerun the analysis without access to the preprocessed data and use different software for model fitting and different random number generation. This allows us to assess if the results are robust to changes in the data retrieval and preprocessing steps as well as the model fitting.
2 Retrieving and Preprocessing the Corpus
In order to reconstruct the corpus web scraping techniques were employed to download the abstracts from the PNAS web page. We ended up with 27,292 abstracts in the period 1991–2001 and with 2,456 in 2001, compared to 28,154 in 1991–2001 and 2,620 in 2001 used by GS2004. This means that we essentially were able to obtain the same number of abstracts. The slight deviations might be due to the fact that our data collection omitted (uncategorized) commentaries, corrections and retractions.
GS2004 did not provide any information if only the abstracts were used or the abstracts combined with the titles. We decided to leave out the paper title for each document because this led to a document-term matrix closer to the one in GS2004. In a first preprocessing step we transformed all characters to lowercase. GS2004 used any delimiting character, including hyphens, to separate words and deleted words which belonged to a standard “stop” list used in computational linguistics, including numbers, individual characters and some function words. We built the document-term matrix with the R [12] package tm [2, 3] and a custom tokenizing function which we deduced from the few exemplary terms in the original paper. Our tokenizer treats non-alphanumeric characters, i.e., characters different from “a”–“z” and “0”–“9”, as delimiters. This step also implicitly strips non-ASCII characters from our downloaded corpus in Unicode encoding, thereby marginally reducing the information in abstracts which contain characters that are widely used in scientific publications, such as those from the Greek alphabet. The minimum word length was set to two and numbers and words in the “stop” list included in package tm were removed. GS2004 further reduced the vocabulary by omitting terms which appeared in less than five documents and we also performed this preprocessing step.
The characteristics of the final document-term matrices are compared in Table 1. Despite the fact that the original set of documents was not the same and a number of preprocessing steps were not clearly specified or slightly differently performed, the final document-term matrices are quite similar with respect to vocabulary size, total occurrence of words and average document length.
3 Model Fitting
GS2004 fit the model using their own software [13]. We use the R package topicmodels [5] with the same settings with respect to number of topics, number of chains, number of samples, length of burn-in interval and sample lag. The implementation of the collapsed Gibbs sampler in the package was written by Xuan-Hieu Phan and coauthors [10].
3.1 Model Selection
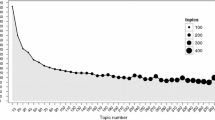
The number of topics are selected by GS2004 using the marginal log-likelihoods determined by the harmonic mean method. Their results are shown in Fig. 3 of their paper and they decide that 300 topics are a suitable choice. For comparison our results are given in Fig. 1. The figure essentially looks quite similar and would lead to the same decision. In the following the topic model fitted with 300 topics is used for further analysis.
3.2 Scientific Topics and Classes
GS2004 used the 33 minor categories which are assigned to each paper to validate whether these class assignments correspond to the differences between the abstracts detected using the statistical analysis method. Using only the abstracts from 2001 we determined the mean topic distribution for each minor category. The most diagnostic topic was then determined as the one where the ratio of the mean value for this category divided by the sum over the mean values of the other categories was greatest. The results are shown in Fig. 2, which corresponds to Fig. 4 in GS2004. Note that our figure includes all 33 minor categories, whereas in the figure in GS2004 category “Statistics” is missing. Again a high resemblance between the two results can be observed. For comparison the five most probable words for the topic assigned to minor category “Ecology” are “species”, “global”, “climate”, “co2” and “water” in GS2004 and “species”, “diversity”, “marine”, “ecological” and “community” in our replication study. This topic also has high mean values for the minor categories “Geology” and “Geophysics” in both solutions.
3.3 Hot and Cold Topics
In a next step GS2004 analyze the dynamics of the topics using a post hoc examination of the mean topic distribution estimates for each year from 1991 to 2001. A linear trend was fitted to each topic over time and the estimated slope parameters were used to identify “hot” and “cold” topics. The five topics with the largest positive and negative slopes in our model are given in Fig. 3. This figure corresponds to Fig. 5 in GS2004 except that they only show the three “hottest” and “coldest” topics. A comparison of results using the twelve most probable words of each topic indicates that matches for the three topics in GS2004 can be identified among the five topics identified by our model, even though the order of the topics is not identical. The coldest topic detected in each of the analyses is remarkably similar, as indicated by a comparison of the twelve most probable words, which are given in Table 2.
3.4 Tagging Abstracts
Each sample of the collapsed Gibbs sampling algorithm consists of a set of assignments of words to topics. These assignments can be used to identify the role words play in documents. In particular this allows to tag each word in the document with the topic to which it was assigned. Our results are given in Fig. 4. The assignments are indicated by the superscripts. Words which do not have a superscript were not included in the vocabulary of the document-term matrix. The shading was determined by averaging over several samples how often the word was assigned to the most prevalent topic of the document. This should be a reasonable estimate even in the presence of label switching. Again a comparison to Fig. 6 in GS2004 indicates that both taggings strongly resemble each other.
Conclusions
The complete analysis presented in GS2004 was reproduced by collecting the data using web scraping techniques, applying preprocessing steps to determine the document-term matrix and fitting the LDA model using collapsed Gibbs sampling. The fitted model was analyzed in the same way as in GS2004: the topic distributions of the minor categories were determined and the most prevalent topics for each minor category are compared with respect to their weight assigned to the minor categories. In addition time trends of the topics were fitted and words in documents were tagged based on the topic assignments from the LDA model. Further results from this replication study and a detailed description of the code used for this analysis are given in [11], except for the use of a slightly different tokenizer. The tokenizer used in [11] is the default in package tm 0.5.1.
Certainly small deviations can be observed between the two results obtained in each of the analyses. However, in general the conclusions drawn as well as the overall assessment are essentially the same. This leads to the conclusion that the study could be successfully reproduced despite the use of completely different tools and a different text database.
Computational Details
For the automated document retrieval from the public PNAS archive (http://www.pnas.org/) we employed Python 2.6.6 with the web scraping framework Scrapy 0.10.3, and additional libraries pycurl 7.19.0-3+b1 and BeautifulSoup 3.1.0.1-2. Texts that were only available as PDF files were converted to plain text with pdftotext 0.12.4.
The main programming and data analysis were conducted in R 2.15.3 with packages tm 0.5-8.3, topicmodels 0.1-9, lattice 0.20-13, xtable 1.7-1 and Rmpfr 0.5-1.
Calculations for model selection and model fitting were delegated to a computer cluster running the Sun Grid Engine at WU (Wirtschaftsuniversität Wien).
References
Blei, D.M., Ng, A.Y., Jordan, M.I.: Latent Dirichlet allocation. J. Mach. Learn. Res. 3, 993–1022 (2003)
Feinerer, I.: tm: Text Mining Package (2013). URL http://CRAN.R-project.org/package=tm. R package version 0.5-8.3
Feinerer, I., Hornik, K., Meyer, D.: Text mining infrastructure in R. J. Stat. Softw. 25(5), 1–54 (2008). URL http://www.jstatsoft.org/v25/i05/
Griffiths, T.L., Steyvers, M.: Finding scientific topics. Proc. Natl. Acad. Sci. U S A 101, 5228–5235 (2004)
Grün, B., Hornik, K.: topicmodels: An R package for fitting topic models. J. Stat. Softw. 40(13), 1–30 (2011). URL http://www.jstatsoft.org/v40/i13/
Hothorn, T., Leisch, F.: Case studies in reproducibility. Brief. Bioinform. 12(3), 288–300 (2011)
Keiding, N.: Reproducible research and the substantive context. Biostatistics 11(3), 376–378 (2010)
Koenker, R., Zeileis, A.: On reproducible econometric research. J. Appl. Econ. 24, 833–847 (2009)
de Leeuw, J.: Reproducible research: the bottom line. Technical Report 2001031101, Department of Statistics Papers, University of California, Los Angeles (2001). URL http://repositories.cdlib.org/uclastat/papers/2001031101/
Phan, X.H., Nguyen, L.M., Horiguchi, S.: Learning to classify short and sparse text & web with hidden topics from large-scale data collections. In: Proceedings of the 17th International World Wide Web Conference (WWW 2008), pp. 91–100. Beijing, China (2008)
Ponweiser, M.: Latent Dirichlet allocation in R. Diploma thesis, Institute for Statistics and Mathematics, WU (Wirtschaftsuniversität Wien), Austria (2012). URL http://epub.wu.ac.at/id/eprint/3558
R Development Core Team: R: A Language and Environment for Statistical Computing. R Foundation for Statistical Computing, Vienna, Austria (2012). URL http://www.R-project.org/. ISBN:3-900051-07-0
Steyvers, M., Griffiths, T.: MATLAB Topic Modeling Toolbox 1.4 (2011). URL http://psiexp.ss.uci.edu/research/programs_data/toolbox.htm
Acknowledgements
This research was supported by the Austrian Science Fund (FWF) under Elise-Richter grant V170-N18.
Author information
Authors and Affiliations
Corresponding author
Editor information
Editors and Affiliations
Rights and permissions
Copyright information
© 2014 Springer-Verlag Berlin Heidelberg
About this chapter
Cite this chapter
Ponweiser, M., Grün, B., Hornik, K. (2014). Finding Scientific Topics Revisited. In: Carpita, M., Brentari, E., Qannari, E. (eds) Advances in Latent Variables. Studies in Theoretical and Applied Statistics(). Springer, Cham. https://doi.org/10.1007/10104_2014_11
Download citation
DOI: https://doi.org/10.1007/10104_2014_11
Published:
Publisher Name: Springer, Cham
Print ISBN: 978-3-319-02966-5
Online ISBN: 978-3-319-02967-2
eBook Packages: Mathematics and StatisticsMathematics and Statistics (R0)