Abstract
The tomato Pto gene encodes a serine/threonine kinase (STK) whose molecular characterization has provided valuable insights into the disease resistance mechanism of tomato and it is considered as a promising candidate for engineering broad-spectrum pathogen resistance in this crop. In this study, a pair of degenerate primers based on conserved subdomains of plant STKs similar to the tomato Pto protein was used to amplify similar sequences in banana. A fragment of ∼550 bp was amplified, cloned and sequenced. The sequence analysis of several clones revealed 13 distinct sequences highly similar to STKs. Based on their significant similarity with the tomato Pto protein (BLASTX E value <3e-53), seven of them were classified as Pto resistance gene candidates (Pto-RGCs). Multiple sequence alignment of the banana Pto-RGC products revealed that these sequences contain several conserved subdomains present in most STKs and also several conserved residues that are crucial for Pto function. Moreover, the phylogenetic analysis showed that the banana Pto-RGCs were clustered with Pto suggesting a common evolutionary origin with this R gene. The Pto-RGCs isolated in this study represent a valuable sequence resource that could assist in the development of disease resistance in banana.
Similar content being viewed by others
Avoid common mistakes on your manuscript.
Introduction
Banana (Musa acuminata) represents a staple food for millions of people living in developing countries and an important export commodity for producing countries (Jain 2004). Banana fruit production is currently threatened by a wide spectrum of pathogens including viruses, bacteria, fungi and nematodes. They reduce yield, affect the appearance, shelf life, and marketability of harvested fruit; debilitate or kill the host plant (Ploetz 2005). Currently, the fungi Mycosphaerella fijiensis and Fusarium oxysporum f. sp. cubense (FOC), which attack the leaves and roots of susceptible bananas, respectively, are the most devastating pathogens of this crop. M. fijiensis is controlled by an intensive regime of fungicide applications which is costly to the growers and detrimental to the environment. The case of FOC is even worse since an effective chemical control does not exist (Ploetz 2005). Genetic resistance is the most suitable strategy to control these pathogens in the field and there are sources of resistance to both of them in Musa germplasm (Jones 2000). However, breeding for pathogen resistance in banana is limited by the long life cycle, triploidy and sterility in most commercial cultivars. Molecular biotechnology has the potential to overcome these constraints by transferring single or even multiple disease resistance (R) genes into the genome of susceptible banana cultivars using either biolistics (Sagi et al. 1995; Becker et al. 2000) or Agrobacterium-mediated transformation (May et al. 1995; Khanna et al. 2004). Although transformation technologies have been developed for banana in various laboratories around the world, no banana R gene has been isolated to date.
Disease resistance in plants occurs in a gene-for-gene manner by a direct or indirect interaction between the plant R gene encoded protein and the pathogen avirulence (Avr) gene encoded protein. Such interactions trigger a cascade of defense responses that halt pathogen spread (Chisholm et al. 2006). These defense responses can also be associated with the development of a localized or, in some cases systemic, programmed cell death known as hypersensitive response (HR) (Gilchrist 1998). R gene-mediated resistance has several attractive features to protect crop plants against pathogen attack. When induced in a timely manner, the concerted responses can efficiently halt pathogen growth with minimal collateral damage to the plant. No input is required from the farmer and there are no adverse environmental effects (McDowell and Woffenden 2003). Consequently, R gene-mediated resistance is one of the top priorities in plant breeding and genetic improvement programs. For effective utilization of these natural R genes and for engineering novel resistance, it is important to clone and characterize them. Over 40 plant R genes that confer resistance to various pathogens including viruses, bacteria, fungi or nematodes have been isolated from several plant species (Martin et al. 2003), using either map-based positional cloning or transposon-tagging. Their gene products have domains consistent with roles in both pathogen recognition and subsequent signal transduction (Dangl and Jones 2001; Hammond-Kosack and Parker 2003). The presence of conserved domains in the protein sequences of these R genes allows their classification into eight classes (Hammond-Kosack and Parker 2003). The tomato Pto gene encodes a cytoplasmic serine/threonine protein kinase (STK) of 321 amino acids and represents one of these classes of R genes. Pto was the first plant R gene cloned that participates in a gene-for-gene interaction with a pathogen (Martin et al. 1993), and it is one of the best-characterized and most intensively studied R genes (Pedley and Martin 2003). Pto confers hypersensitive response-mediated resistance against strains of Pseudomonas syringae pv. tomato that express the avirulence proteins AvrPto or AvrPtoB (Martin et al. 1993; Kim et al. 2002). Overexpression of Pto in tomato under the control of the strong cauliflower mosaic virus (CaMV) 35S promoter has been shown to activate defense responses in the absence of pathogen inoculation. Pto-overexpressing plants show resistance not only to P. syringae pv tomato but also to Xanthomonas campestris pv vesicatoria and to the fungal pathogen Cladosporium fulvum (Tang et al. 1999). These findings make Pto an interesting candidate for engineering broad-spectrum pathogen resistance in agriculture and encourage the search for functional Pto-like genes in other plant species.
Degenerate primers designed from conserved motifs of the nucleotide binding site-leucine rich repeat (NBS-LRR) class of R genes have been used to amplify R-like genes or resistance gene candidates (RGCs) from genomic DNA of numerous plants (Collins et al. 1998; Noir et al. 2001; López et al. 2003; Martínez-Zamora et al. 2004). Many of these RGCs are phylogenetically related with known R genes (Meyers et al. 1999) and some of them have facilitated the cloning of full-length functional R genes from different plant species (McDowell et al. 1998; Zhao et al. 2005). In addition to their potential use for genetic improvement, RGCs also provide opportunities and tools to answer some fundamental questions about disease resistance genes, such as structure, R gene organization, distribution and evolution (Michelmore and Meyers 1998; Meyers et al. 1999). The use of PCR with degenerate primers targeting the highly conserved subdomains of STK proteins has also proven to be an efficient method for isolating Pto resistance gene candidates (Pto-RGCs) in bean and grapevine (Vallad et al. 2001; Di Gaspero and Cipriani 2003), indicating that this approach could be used to retrieve this type of gene from other plant species. Since no molecular characterization of banana Pto-RGCs has been published, the objectives of this study were: (1) to obtain Pto-RGCs from banana using degenerate PCR and (2) to determine the structure and phylogenetic relationships of the banana Pto-RGCs.
Materials and methods
Plant material and DNA extraction
The banana cultivar ‘Tuu Gia’ was chosen for PCR amplification of Pto-RGC sequences because it shows resistance to a range of banana pathogens, including the most destructive such as M. fijiensis and F. oxysporum, and also because there is a binary bacterial artificial chromosome (BIBAC) genomic library from this cultivar available for banana transformation (Ortiz-Vázquez et al. 2005). The plant material was obtained from the international network for the improvement of banana and plantain (INIBAP) Musa Germplasm transit centre in Leuven, Belgium. The youngest fully developed leaf from a greenhouse-grown ex vitro plant of Musa acuminata cv. ‘Tuu gia’ (accession no. ITC 0610) was harvested for nucleic acid extraction. The genomic DNA was isolated using the Nucleon PhytoPure® kit (Amersham™) according to manufacturer’s instructions.
Degenerate PCR
A pair of degenerate primers designed by Vallad et al. (2001), forward 5′-TNGGNSANGGNGKNTTYGG-3′ and reverse 5′-ACNCCRAANGARTANACRTC-3′, was used to amplify the region between the subdomains I and IX of STKs. The degenerate PCR reaction was performed in a 50-μl reaction volume containing 300 μM of dNTPs, 4 μM of each degenerate primer forward and reverse, 1 U of Taq DNA polymerase (Invitrogen™), 1× PCR buffer, 1.5 mM MgCl2 and approximately 200 ng of genomic DNA. PCR conditions were 95°C for 3 min, followed by 35 cycles of 95°C for 30 s, 45°C for 30 s and 72°C for 1 min; and an additional 10 min extension at 72°C was included.
Cloning and sequencing
PCR products were visualized on a 1% agarose gel stained with ethidium bromide. A band of the expected size was excised from the gel and purified using the High Pure Purification kit (Roche) according to manufacturer’s instructions. Purified PCR products were cloned into the pGEM-T easy plasmid vector (Promega). Plasmids were transferred by electroporation into Escherichia coli DH10B competent cells. Bacteria were plated onto LB medium containing ampicillin, X-Gal and IPTG, and recombinant plasmids were chosen by blue/white selection (Sambrook and Russell 2001). Plasmid DNA was purified by the alkaline lysis method (Sambrook and Russell 2001) and sequenced using the BigDye terminator sequencing kit version 3.1 (Applied Biosystems) according to manufacturer’s instructions. The sequencing products were separated with an ABI 3730×1 automatic sequencer (Applied Biosystems) through the capillary separation service of the Australian Genome Research Facility (AGRF) (http://www.agrf.org.au). Selected clones were sequenced in both orientations.
Sequence edition, similarity searches and multiple sequence alignment
All sequences were assembled and edited using the programs SEQMAN and EDIT, respectively of the Lasergene software package version 4.03 (DNASTAR, Madison, WI, USA). The degenerate primer sequences were removed from each sequenced clone so only the region between the end of subdomain I and the start of subdomain IX of STKs was considered for further analysis. Predicted amino acid sequences were generated using the translate tool of the EDIT program (Lasergene software). Similarity searches were conducted with the BLASTX program (Altschul et al. 1997) through the National Center for Biotechnology Information (NCBI) GenBank database (http://www.ncbi.nlm.nih.gov) using the default settings. Percent amino acid identity between predicted protein sequences was determined with the MEGALIGN program of the Lasergene software using the default settings. Determination of conserved amino acids in banana Pto-RGC sequences was carried out with the programs ClustalX version 1.81 (Thompson et al. 1997) and WebLogo version 2.8.2 (Crooks et al. 2004)(http://weblogo.berkeley.edu/) using the default settings.
Phylogenetic analysis
Phylogenetic trees were constructed by the neighbor-joining (NJ) method (Saitou and Nei 1987) using the NJ algorithm implemented in the Molecular Evolutionary Genetics Analysis (MEGA) software version 2.1 (Kumar et al. 2001) with the Poisson correction. Bootstrapping (1,000 replicates) was used to evaluate the degree of support for a particular grouping pattern in the phylogenetic tree. Protein sequences belonging to 12 groups of characterized STKs from Arabidopsis thaliana (Hardie 1999), a phosphoenolpyruvate carboxylase kinase (PEPck) (GenBank accession no. AF162660) from A. thaliana (Hartwell et al. 1999), the tomato Pto protein (GenBank accession no. A49332) and Pto-RGCs from different plant species were retrieved from the GenBank for the phylogenetic tree constructions. The tomato Pto disease resistant protein was used as query in BLASTP (Altschul et al. 1997) searches to retrieve amino acid sequences of Pto-RGCs from the GenBank. Only the region between the end of subdomain I and the start of subdomain IX was considered for the phylogenetic tree constructions.
Results
Identification of Pto resistance gene candidates in banana
PCR amplification of banana genomic DNA with a pair of degenerate primers previously used by Vallad et al. (2001) generated an expected band of ∼550 bp. This band was cloned and a total of 70 clones were sequenced. The primer sequences were removed from each sequenced clone for further analysis. Of the 70 sequenced clones (Tg-1 to Tg-70), 56 presented uninterrupted open reading frames (ORFs), while the other 14 sequences presented multiple stop codons in all reading frames, and as a result they were not further investigated. Similarity searches of the 56 banana sequences using the BLASTX algorithm (Altschul et al. 1997) against the NCBI non-redundant database revealed significant similarity to known STKs (E value <3e-53), including the disease resistance protein Pto from tomato. A threshold value of 85% amino acid identity previously used by Vallad et al. (2001) to classify Pto-RGC clones from bean into classes or groups was used in the present study, therefore banana clones with greater than 85% amino acid identity were considered to be part of the same group. A total of 13 distinct groups of STK-like sequences were identified, most of which contained redundant or highly similar clones (>97% amino acid identity). Seven groups were designated as Pto resistance gene candidates (Pto-RGCs) based on their significant similarity with the tomato Pto disease resistance protein (E value <3e-53). The other six groups showed significant similarity to other types of STKs, which are described later in the text. Each group was designated by the name of a single clone representative of the group and used for further analysis. Percent amino acid identity between the predicted amino acid sequence of Pto-RGCs and the corresponding region of the Pto protein ranged from 58% (Tg-67) to 68% (Tg-13) (Table 1), whereas amino acid identity among the Pto-RGCs ranged from 84.1% (Tg-4 vs. Tg-13) to 65.3% (Tg-10 vs. Tg-12) (Table 1). BLASTX searches also revealed that two Pto-RGCs (Tg-9 and Tg-12) were highly similar (>92% amino acid identity) to Musa sequences present in the GenBank database as accession nos. AAM97913 and AAM97914 from Musa balbisiana and Musa acuminata, respectively. These sequences were considered to be part of the groups represented by the sequences Tg-12 and Tg-9, respectively.
Isolation of other banana serine/threonine kinase-like sequences
The degenerate primers used in this study were designed from the conserved subdomains I and IX of the STKs Pto, Fen and Pti1 of tomato, and MHK and APK1 of Arabidopsis (Vallad et al. 2001). Therefore, these primers have the potential to isolate not only Pto-RGCs but also other types of plant STKs. In agreement with this observation six additional STK-like sequences from banana were identified in BLASTX searches (Table 2). Five of them (Tg-1, Tg-20, Tg-34, Tg-36 and Tg-55) showed significant similarity to the receptor-like kinase (RLK) subfamily (E value <2e-64), whereas the remaining sequence Tg-2 showed a significant similarity to a putative PEPck (E value = 8e-74) from Oryza sativa.
Multiple sequence alignment and phylogenetic analysis
A multiple sequence alignment using the ClustalX program was performed with the predicted amino acid sequences of the seven banana Pto-RGCs and the corresponding region of the tomato Pto protein (Fig. 1). The alignment revealed that several features of the Pto protein are highly conserved in the banana Pto-RGCs such as the STK subdomains internal to the degenerate primer sequences, the presence of the activation domain between subdomains VII and VIII, and its internal P+1 loop site, which is responsible for the specific binding of AvrPto (Frederick et al. 1998), and also several invariant amino acids distributed throughout the sequences. In addition, three of the four autophosphorylation sites (serine or threonine) in the activation domain of Pto (Sessa et al. 2000) are conserved in the corresponding region of all banana Pto-RGCs (Fig. 1). The alignment also showed that all the banana Pto-RGCs presented a two amino acid deletion (subdomain V) and a three amino acid insertion (subdomain VIa) with regard to the Pto protein. We found that the two amino acid deletion was also present in Pto-RGCs from other monocot species such as Oryza sativa (GenBank accession no. XP476621) and Triticum aestivum (GenBank accession no. AAL51075). This deletion was also present in Pto-RGCs from other dicot species such as Arabidopsis thaliana (GenBank accession no. NP197789) and Cucumis sativus (GenBank accession no. AAP57674) but absent in Phaseolus vulgaris (GenBank accession no. AF363819). The extent and significance of this polymorphism in both monocot and dicot Pto-RGCs awaits further research. In the case of the three amino acid insertion, it is present in Pto-RGCs from other monocot and dicot species but absent in Pto-RGCs from the Solanaceae family (Vleeshouwers et al. 2001).
ClustalX alignment of the deduced amino acid sequences of seven banana Pto-RGC sequences together with the corresponding region of the Pto protein. The subdomains, activation domain and P + 1 loop of protein kinases (Hanks et al. 1988; Hanks and Quinn 1991; Vallad et al. 2001) are indicated. Conserved amino acids in plant serine/threonine kinases (Hanks and Quinn 1991) and Pto autophosphorylation sites (Sessa et al. 2000) are indicated with asterisks and grey circles, respectively. Identical amino acids are shaded in black and conservative substitutions are shaded in grey
In order to highlight the Pto autophosphorylation sites that are conserved in the banana Pto-RGCs and other critical residues for Pto function located in the activation domain, a sequence Logo was generated with the banana Pto-RGC products and it is shown in Fig 2. Of the three Pto autophosphorylation sites (Thr195, Ser198 and Thr199) conserved in the banana Pto-RGCs, Ser198 is required for the AvrPto-Pto-mediated hypersensitive response (Sessa et al. 2000) and it is present in the majority of banana Pto-RGCs with the exception of Tg-12, in which the serine has been replaced by threonine, thereby providing an alternative phosphorylation site. The Pto (Thr190) autophosphorylation site was replaced by a proline in the banana Pto-RGCs with the exception of Tg-6, which contains a threonine at this site. Two other Pto residues crucial for Pto function but not autophosphorylated in vitro are Thr204 and Tyr207 (Pedley and Martin 2003). Individual mutations in these residues (pto Thr204Ala or pto Tyr207Ala) abolished Pto interaction with AvrPto in yeast and transient expression of the mutants pto Thr204Asp and pto Tyr207Asp in leaf tissue of Nicotiana benthamiana produced a hypersensitive response-like cell death in the absence of AvrPto (the phenotype of pto Tyr207Aspwas the stronger of the two mutant genes) (Rathjen et al. 1999). The sequence Logo showed that Pto (Thr204) was replaced by a serine in the majority of banana Pto-RGCs and Pto (Tyr207) was present in all Pto-RGCs (Fig. 2).
Comparison of the tomato Pto activation domain (30 amino acids in length) with the sequence Logo of the putative activation domain of seven banana Pto-RGCs. The Logo sequence consists of stacks of letters, one stack for each position in the sequence. The overall height of each stack indicates the sequence conservation at that position (measured in bits), whereas the height of symbols within the stack reflects the relative frequency of the corresponding amino acid at that position (Crooks et al. 2004). The autophosphorylation sites in the activation domain of Pto (Sessa et al. 2000) are indicated with grey circles
The phylogenetic analysis of Fig. 3 shows that the seven banana STK-like sequences identified as Pto-RGCs formed a cluster with the tomato Pto protein, which is supported by a high bootstrap value (99%). This result supported the designation of the seven banana STK-like sequences as Pto-RGCs. The most closely related sequences to the Pto-like kinase cluster were sequences of the receptor-like kinase subfamily (Hardie 1999). Regarding the other banana STK-like sequences, five of them were related to receptor-like kinases as previously observed in the BLASTX results and the remaining Tg-2 sequence formed a highly supported cluster with a PEPck of Arabidopsis (Hartwell et al. 1999) (Fig. 3). This phylogenetic tree also showed that the protein kinase region used for its construction contains sufficient sequence information to represent clusters defined by analysis with full sequence data of the kinase catalytic domain (Hardie 1999). Of the 12 groups of plant STKs (Hardie 1999) used for the phylogenetic analysis, only the receptor-like kinases were not grouped in a single cluster, which is consistent with the phylogenetic tree performed by Vallad et al. (2001). Furthermore, phylogenetic analysis of the banana Pto-RGCs with Pto-RGCs from different plant species (Fig. 4) revealed that the banana Pto-RGCs were more closely related to Pto-RGCs from other plant species than each other. Another interesting finding was that the clades where the banana Pto-RGCs were grouped (I, II and III) also contain Pto-RGCs from dicot species, suggesting that the origin of this type of sequence may have preceded the divergence of monocot and dicot plants.
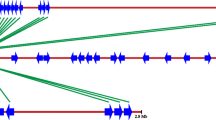
Neighbor-joining phylogenetic tree based on the ClustalX alignment of serine/threonine kinases from Arabidopsis thaliana (Hardie 1999; Hartwell et al. 1999), tomato Pto disease resistance protein, Pto-RGCs (black circles) and other STK-like sequences (grey circles) from banana. Names of STKs are given for each sequence followed by GenBank accession numbers in parentheses. Amino acids from the end of subdomain I to the start of subdomain IX of STKs were used for the analysis. The numbers above the branches indicate the percentage of 1,000 bootstrap replications supporting the particular nodes, and only those with >60% support are shown
Neighbor-joining phylogenetic tree based on the ClustalX alignment of tomato Pto disease resistance protein, Pto-RGCs from banana (black circles) and other Pto-RGCs from monocot and dicot species. The name of the Pto protein is in bold and GenBank accession numbers are given for each sequence followed by species name in italics. Amino acids from the end of subdomain I to the start of subdomain IX of STKs were used for the analysis. The numbers above the branches indicate the percentage of 1,000 bootstrap replications supporting the particular nodes, and only those with >60% support are shown
Discussion
There is evidence that Pto-RGCs are highly conserved in many plant species. Southern hybridization using the tomato Pto gene as probe revealed the presence of Pto-RGCs in many plant species such as Arabidopsis, bean, soybean, pea, rice, maize, barley, wheat and sugarcane (Martin et al. 1993). Recent studies report the cloning and characterization of Pto-RGCs from potato, bean and grapevine (Vleeshouwers et al. 2001; Vallad et al. 2001; Di Gaspero and Cipriani 2003). Furthermore, other Pto-RGC sequences from different plant families have been deposited in the GenBank (http://www.ncbi.nlm.nih.gov), however these sequences have not been characterized. In this study, a set of Pto-RGC sequences and other STK-like sequences were identified from the banana cultivar ‘Tuu Gia’. These sequences were isolated by PCR using a pair of degenerate primers previously designed and used by Vallad et al. (2001) to isolate Pto-RGCs from bean. In total, seven distinct Pto-RGC sequences and six other STK-like sequences were identified in the banana genome. Vallad et al. (2001) reported the identification of a lower number of Pto-RGCs (five distinct sequences sharing from 56.9 to 63.9% amino acid identity with Pto) and no further cloning of other STK-like sequences. The PCR annealing temperature used in this previous study was high (60°C) in comparison to our study that used a less stringent temperature of 45°C, which may explain the broader diversity of STK-like sequences isolated in banana. This low PCR annealing temperature could explain the isolation of the banana PEPck-like sequence, which was quite divergent from the rest of the banana STK-like sequences isolated (Pto-RGCs and RLKs) (Fig. 3). Although, the degenerate primers amplified preferentially Pto-RGCs and RLK sequences, we believe that they could still be exploited to isolate other types of STK-like sequences by lowering the annealing temperature of the PCR. Overall, our data demonstrated that the degenerate primers used are capable of amplifying Pto-RGCs and other types of STK-like sequences from a monocot species.
The complete genome sequence of Arabidopsis (genome size of 130 Mbp) revealed the presence of 15 Pto-RGCs (Arabidopsis Genome Initiative 2000), while a draft of the rice genome sequence (genome size of 420 Mbp) revealed a similar number of Pto-RGCs with 14 (Goff et al. 2002). These data indicate that the number of Pto-RGCs in these two plant genomes is conserved even though the rice genome is 290 Mbp larger than Arabidopsis, and also indicate that the number of Pto-RGCs in a plant genome is small in comparison to the NBS-LRR class of R genes, which has a large number of divergent genes in the Arabidopsis and rice genomes, with 149 and 480 genes, respectively (Meyers et al. 2003; Zhou et al. 2004). The genome size of Musa acuminata is estimated to be 600 Mbp (Dolezel et al. 1994), assuming that the number of Pto-RGCs in a plant genome do not increase significantly according to the genome size, then it is possible that in banana the number of Pto-RGCs could be similar to Arabidopsis or rice. Hence, it is tempting to speculate that the number of Pto-RGC sequences identified in this study represents a significant proportion of the total number of Pto-RGC sequences in the banana genome. Indeed, the complete sequence of the banana genome will provide comprehensive data on the number of Pto-RGC sequences in this Musaceae species (http://www.musagenomics.org).
All banana Pto-RGC products displayed conserved serine-threonine kinase subdomains (Hanks and Quinn 1991), suggesting that the uncovered genes are likely to encode active kinases. Moreover, most residues of the Pto activation domain involved in pathogen recognition and HR induction (Pedley and Martin 2003) are highly conserved in banana Pto-RGCs suggesting that these residues might play a similar role in banana. Indeed, the cloning of the full cDNA sequence and protein expression of these banana Pto-RGCs will allow the possibility to answer some fundamental questions regarding for example, whether the Pto-RGC encoded proteins are autophosphorylated in vitro and also whether substitution of tyrosine by aspartate in the corresponding site of Pto (Tyr207) will lead to a HR-like induction. Regarding the other STK-like sequences reported in this study, some of them are similar to receptor-like kinases that are known to be involved in the response to pathogens, for example banana Tg-1 was related to the cell wall-associated kinase (WAK) group. WAK1, a member of this kinase group in Arabidopsis, is required for plants to survive P. syringae infection. Moreover, WAK1 expression is induced by salicylic acid in an NPR1-dependent manner, demonstrating that it is a pathogenesis-related gene (He et al. 1998). Other interesting examples are Tg-20 and Tg-36, which were related to the Lectin receptor-like kinase (LecRLK) group. A LecRLK gene is strongly induced upon chitin elicitation in Arabidopsis (Zhang et al. 2002), and recently, a LecRLK gene from rice (Pi-d2) was cloned and identified as a novel class of R gene involved in conferring resistance to Magnaporthe grisea (Chen et al. 2006). The role of the banana RLK sequences in disease resistance remains to be determined.
Phylogenetic analyses of Pto and Pto-RGC sequences have suggested that these sequences form a unique group of kinases in plants (Vallad et al. 2001; Vleeshouwers et al. 2001). In agreement with this finding the banana Pto-RGCs formed a highly supported group with the Pto disease resistance protein (Fig. 3) suggesting that these sequences share a common evolutionary origin with the tomato Pto protein and possibly a similar function in disease resistance. Furthermore, phylogenetic analysis of Pto-RGCs from different Solanum species has revealed that Pto orthologue genes are more similar than paralogues suggesting that the origin of Pto could predate the radiation of Solanum species (Vleeshouwers et al. 2001). This ancient origin of Pto is further supported by the fact that both Pto and a Pto orthologue (LhirPto) are functional in Nicotiana benthamiana (Riely and Martin 2001). Additional evidence of this ancient origin is the presence of Pto-RGCs in other dicot species and also monocots that have been recently deposited in the GenBank (http://www.ncbi.nlm.nih.gov). The phylogenetic analysis of Fig. 4 supports and extends these previous observations since all banana Pto-RGCs were grouped in clades that contained Pto-RGCs from both monocot and dicot species suggesting that the origin of this type of sequence might have predated the divergence of monocot and dicot plants which took place about 200 ± 40 million years ago (Wolfe et al. 1989).
The tomato Pto protein is capable of recognizing at least two Avr proteins (AvrPto and AvrPtoB) from P. syringae (Kim et al. 2002). Surprisingly, these two Avr proteins share limited sequence similarity. This dual recognition specificity has also been reported in R proteins of the NBS-LRR class, for example the Rpm1 protein from Arabidopsis confers resistance to P. syringae and recognizes two different avirulence proteins, AvrB and AvrRpm1 (Bisgrove et al. 1994). Another interesting example is the Mi-1 gene from tomato, which confers resistance to a nematode and an aphid pest (Vos et al. 1998). This dual (and perhaps even multiple) pathogen recognition specificity for a single R protein may prove to be common in R genes (Martin et al. 2003) and raises the possibility that Pto may confer resistance to pathogens other than bacteria. Whether the banana Pto-RGCs are involved in conferring bacterial resistance as in tomato or are involved in conferring resistance to other types of pathogens will require functional analysis, which could be carried out with genetic complementation or loss-of-function experiments. In the case of genetic complementation, the banana Pto-RGCs could be used as probes to screen a banana BIBAC library (Ortiz-Vázquez et al. 2005) for the isolation of BIBAC clones containing Pto-RGCs. These Pto-RGC-BIBAC clones could be used to transform a banana disease susceptible cultivar using Agrobacterium tumefaciens (Khanna et al. 2004). These experiments would lead to a collection of Pto-RGC-BIBAC transgenic lines ready to be used for disease resistance tests. The BIBAC technology coupled with Agrobacterium-mediated transformation not only promises to unravel the function of banana RGCs but also the development of disease resistance in this crop. In the case of the loss-of-function strategy, the banana Pto-RGC sequences could be used in RNA interference (RNAi) constructs (Waterhouse and Helliwell 2003) in order to silence their corresponding targets in a resistant genotype. Those resistant plants that show disease symptoms after the infection with a particular type of pathogen would allow the identification of an R gene. The RNAi technology has been recently used to determine the function of genes involved in disease resistance in barley (Douchkov et al. 2005). The banana Pto-RGCs could also be used to produce molecular markers tightly linked to R genes for genomic mapping and positional cloning. In this respect, several RGCs of the NBS-LRR class have been shown to be quite useful as molecular markers to assist the isolation of functional R genes through map-based positional cloning (McDowell et al. 1998; Zhao et al. 2005).
The Pto gene is considered as a promising candidate for engineering broad-spectrum pathogen resistance in tomato since plants overexpressing this gene display resistance to both bacterial and fungal pathogens (Tang et al. 1999). Moreover, expression of Pto mutants such as pto Thr204Asp or pto Tyr207Asp can constitutively activate a HR-like response in the absence of P. syringae (Rathjen et al. 1999). Expression of these engineered Pto genes under the control of a defined inducible promoter has been considered as another promising strategy to protect crops against pathogens through the hypersensitive response (Rathjen et al. 1999). The cloning of the full cDNA sequences of the banana Pto-RGCs will permit assessing their potential to confer disease resistance using the strategies mentioned above.
In summary, this study has uncovered a set of banana Pto-RGC sequences and provided the first insights about their amino acid sequence structure and evolution. The presence of several conserved amino acids in the banana Pto-RGCs that are crucial for Pto function, and the fact that these sequences were phylogenetically closely related to Pto, make of them a valuable sequence resource for plant–pathogen interaction studies in banana. The banana Pto-RGCs could be used to generate not only a collection of BIBAC clones or RNAi constructs for functional analysis but also they might be useful as molecular markers for genetic mapping. The availability of these sequences will facilitate the cloning of their corresponding full gene sequences, which in turn will allow further genetic and biochemical characterization that may lead to the development of specific or even broad-spectrum pathogen resistance in banana. Moreover, the other banana STK-like sequences identified in this study may be used as a research platform for further studies in this crop.
References
Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402
Arabidopsis Genome Initiative (2000) Analysis of the genome sequence of the flowering plant Arabidopsis thaliana. Nature 408:796–815
Becker D, Dugdale B, Smith M, Harding R, Dale J (2000) Genetic transformation of Cavendish banana (Musa spp. AAA group) cv ‘Grand Nain’ via microprojectile bombardment. Plant Cell Rep 19:229–234
Bisgrove SR, Simonich MT, Smith NM, Sattler A, Innes RW (1994) A disease resistance gene in Arabidopsis with specificity for two different pathogen avirulence genes. Plant Cell 6:927–933
Chen X, Shang J, Chen D, Lei C, Zou Y, Zhai W, Liu G, Xu J, Ling Z, Ma B, Wang Y, Zhao X, Li S, Zhu L (2006) A B-lectin receptor kinase gene conferring rice blast resistance. Plant J 46:794–804
Chisholm ST, Coaker G, Day B, Staskawicz BJ (2006) Host–microbe interactions: shaping the evolution of the plant immune response. Cell 124:803–814
Collins NC, Webb CA, Seah S, Ellis JG, Hulbert SH, Pryor A (1998) The isolation and mapping of disease resistance gene analogs in maize. Mol Plant Microbe Interact 11:968–978
Crooks GE, Hon G, Chandonia JM, Brenner E (2004) WebLogo: a sequence Logo generator. Genome Res 14:1188–1190
Dangl JL, Jones JDG (2001) Plant pathogens and integrated defense responses to infection. Nature 411: 826–833
Di Gaspero G, Cipriani G (2003) Nucleotide binding site/leucine-rich repeats, Pto-like and receptor-like kinases related to disease resistance in grapevine. Mol Genet Genomics 269:612–623
Dolezel J, Dolezelova M, Novak FJ (1994) Flow cytometric estimation of nuclear DNA amount in diploid bananas (Musa acuminata and M. balbisiana). Biol Plant 36:351–357
Douchkov D, Nowara D, Zierold U, Schweizer (2005) A high-throughput gene-silencing system for the functional assessment of defense-related genes in barley epidermal cells. Mol Plant Microbe Interact 18:755–761
Frederick RD, Thilmony RL, Sessa G, Martin GB (1998) Recognition specificity for the bacterial avirulence protein AvrPto is determined by Thr-204 in the activation loop of the tomato Pto kinase. Mol Cell 2:241–245
Gilchrist D.G. (1998) Programmed cell death in plant disease: the purpose and promise of cellular suicide. Annu Rev Phytopathol 36:393–414
Goff SA, Ricke D, Lan TH, Presting G, Wang R, Dunn M, Glazebrook J, Sessions A, Oeller P, Varma H, Hadley D, Hutchison D, Martin C, Katagiri F, Lange BM, Moughamer T, Xia Y, Budworth P, Zhong J, Miguel T, Paszkowski U, Zhang S, Colbert M, Sun WL, Chen L, Cooper B, Park S, Wood TC, Mao L, Quail P, Wing R, Dean R, Yu Y, Zharkikh A, Shen R, Sahasrabudhe S, Thomas A, Cannings R, Gutin A, Pruss D, Reid J, Tavtigian S, Mitchell J, Eldredge G, Scholl T, Miller RM, Bhatnagar S, Adey N, Rubano T, Tusneem N, Robinson R, Feldhaus J, Macalma T, Oliphant A, Briggs S (2002) A draft sequence of the rice genome (Oryza sativa L. ssp. japonica). Science 296:92–100
Hammond-Kosack K, Parker J (2003) Deciphering plant-pathogen communication: fresh perspectives for molecular resistance breeding. Curr Opin Biotech 14:177–193
Hanks SK, Quinn AM, Hunter T (1988) The protein kinase family: conserved features and deduced phylogeny of the catalytic domains. Science 241:42–52
Hanks SK, Quinn AM (1991) Protein kinase catalytic domain sequence database: identification of conserved features of primary structure and classification of family members. Method Enzymol 200:38–62
Hardie DG (1999) Plant protein serine/threonine kinases: classifications and functions. Annu Rev Plant Physiol Plant Mol Biol 50:97–131
Hartwell J, Gill A, Nimmo GA, Wilkins MB, Jenkins GI, Nimmo HG (1999) Phosphoenolpyruvate carboxylase kinase is a novel protein kinase regulated at the level of expression. Plant J 20:333–342
He ZH, He D, Kohorn BD (1998) Requirement for the induced expression of a cell wall associated receptor kinase for survival during the pathogen response. Plant J 14:55–63
Jain SM (2004) Cellular biology and biotechnology including mutation techniques for creation of new useful banana genotypes. In: Jain SM, Swennen R (eds) Banana improvement: cellular, molecular biology, and induced mutations. Science Publishers, USA
Jones DR (2000) History of banana breeding. In: Jones DR (eds) Diseases of Banana, Abacá and Enset. CABI, UK, pp 425–434
Khanna H, Becker D, Kleidon J, Dale J (2004) Centrifugation assisted Agrobacterium-mediated transformation (CAAT) of embryogenic cell suspensions of banana (Musa spp. Cavendish AAA and Lady finger AAB). Mol Breeding 14:239–252
Kim YJ, Lin NC, Martin GB (2002) Two distinct Pseudomonas effector proteins interact with the Pto kinase and activate plant immunity. Cell 109:589–598
Kumar S, Tamura K, Jakobsen IB, Nei M (2001) MEGA2: Molecular evolutionary genetics analysis software. Bioinformatics 17:1244–1245
López C, Acosta I, Jara C, Pedraza F, Gaitan-Solis E, Gallego G, Beebe S, Tohme J (2003) Identifying resistance gene analogs associated with resistance to different pathogens in common bean. Phytopathology 93:88–95
Martin GB, Brommonschenkel SH, Chunwongse J, Frary A, Ganal MW, Spivey R, Wu T, Earle ED, Tanksley SD (1993) Map-based cloning of a protein kinase gene conferring disease resistance in tomato. Science 262:1432–1435
Martin GB, Bogdanove AJ, Sessa G (2003) Understanding the function of plant disease resistance proteins. Annu Rev Plant Biol 54:23–61
Martínez-Zamora MG, Castagnaro AP, Diaz-Ricci JC (2004) Isolation and diversity analysis of resistance gene analogues (RGAs) from cultivated and wild strawberries. Mol Genet Genomics 272:480–487
May G, Afza R, Mason H, Wiecko A, Novak F, Arntzen C (1995) Generation of transgenic banana (Musa acuminata) plants via Agrobacterium-mediated transformation. Bio/Technology 13:486–492
McDowell JM, Dhandaydham M, Long TA, Aarts MGM, Goff S, Holub EB, Dangl JL (1998) Intragenic recombination and diversifying selection contribute to the evolution of downy mildew resistance at the RPP8 locus of Arabidopsis. Plant Cell 10:1861–1874
McDowell JM, Woffenden BJ (2003) Plant disease resistance genes: recent insights and potential applications. Trends Biotech 21:178–183
Meyers BC, Dickerman AW, Michelmore RW, Sivaramakrishnan S, Sobral BW, Young ND (1999) Plant disease resistance genes encode members of an ancient and diverse protein family within the nucleotide-binding superfamily. Plant J 20:317–332
Meyers BC, Kozik A, Griego A, Kuang H, Michelmore RW (2003) Genome-wide analysis of NBS-LRR-encoding genes in Arabidopsis. Plant Cell 15:809–834
Michelmore RW, Meyers BC (1998) Clusters of resistance genes in plants evolve by divergent selection and a birth-and-death process. Genome Res 8:1113–1130
Noir S, Combes MC, Anthony F, Lashermes P (2001) Origin, diversity and evolution of NBS-type disease resistance gene homologues in coffee trees (Coffea L.). Mol Genet Genomics 265:654–662
Ortiz-Vázquez E, Kaemmer D, Zhang HB, Muth J, Rodríguez-Mendiola M, Arias-Castro C, James A (2005) Construction and characterization of a plant transformation-competent BIBAC library of the black Sigatoka resistant banana Musa acuminata cv. Tuu Gia (AA). Theor Appl Genet 110:706–713
Pedley KF, Martin GB (2003) Molecular basis of Pto-mediated resistance to bacterial speck disease in tomato. Annu Rev Phytopathol 41:215–243
Ploetz RC (2005) Panama disease, an old nemesis rears its ugly head: part 1, the beginnings of the banana export trades. Plant Health Progress. doi:10.1094/PHP-2005-1221-01-RV
Rathjen JP, Chang JH, Staskawicz BJ, Michelmore RW (1999) Constitutively active Pto induces a Prf-dependent hypersensitive response in the absence of avrPto. EMBO J 18:3232–3240
Riely BK, Martin GB (2001) Ancient origin of pathogen recognition specificity conferred by the tomato disease resistance gene Pto. Proc Natl Acad Sci USA 98:2059–2064
Sagi L, Panis B, Remy S, Schoofs H, De Smet K, Swennen R, Cammue P (1995) Genetic transformation of banana and plantain (Musa spp) via particle bombardment. Bio/Techniques 13:481–485
Saitou N, Nei M (1987) The neighbor-joining method: a new method for reconstructing phylogenetic trees. Mol Biol Evol 4:406–425
Sambrook J, Russell D (2001) Molecular cloning: a laboratory manual, 3rd edn. Cold Spring Harbor Laboratory Press, Cold Spring Harbor
Sessa G, D’Ascenzo M, Martin GB (2000) Thr38 and Ser198 are Pto autophosphorylation sites required for the AvrPto-Pto-mediated hypersensitive response. EMBO J 19:2257–2269
Tang X, Xie M, Kim YJ, Zhou J, Klessig DF, Martin GB (1999) Overexpression of Pto activates defense responses and confers broad resistance. Plant Cell 11:15–29
Thompson JD, Gibson TJ, Plewniak F, Jeanmougin F, Higgins DG (1997) The ClustalX windows interface: flexible strategies for multiple sequence alignment aided by quality analysis tools. Nucleic Acids Res 24:4876–4882
Vallad G, Rivkin M, Vallejos C, McClean P (2001) Cloning and homology modelling of a Pto-like protein kinase family of common bean (Phaseolus vulgaris L.). Theor Appl Genet 103:1046–1058
Vleeshouwers V.G.A.A, Martens A, van Dooijeweert W, Colon LT, Govers F, Kamoun S (2001) Ancient diversification of the Pto kinase family preceded speciation in Solanum. Mol Plant Microbe Interact 14:996–1005
Vos P, Simons G, Jesse T, Wijbrandi J, Heinen L, Hogers R, Frijters A, Groenendijk J, Diergaarde P, Reijans M, Fierens-Onstenk J, Both M, Peleman J, Liharska T, Hontelez J, Zabeau M (1998) The tomato Mi-1 gene confers resistance to both root-knot nematodes and potato aphids. Nature Biotech 16:1365–1369
Waterhouse PM, Helliwell CA (2003) Exploring plant genomes by RNA-induced gene silencing. Nature Rev 4:29–38
Wolfe KH, Gouy M, Yang YW, Sharp PM, Li WH (1989) Date of the monocot-dicot divergence estimated from chloroplast DNA sequence data. Proc Natl Acad Sci USA 86:6201–6205
Zhang B, Ramonell K, Somerville S, Stacey G (2002) Characterization of early, chitin-induced gene expression in Arabidopsis. Mol Plant Microbe Interact 15:963–970
Zhao B, Lin X, Poland J, Trick H, Leach J, Hulbert S (2005) A maize resistance gene functions against bacterial streak disease in rice. Proc Natl Acad Sci USA 102:15383–15388
Zhou T, Wang T, Chen JQ, Araki H, Jing Z, Jiang K, Shen J, Tian D (2004) Genome-wide identification of NBS genes in japonica rice reveals significant expansion of divergent non-TIR NBS-LRR genes. Mol Genet Genomics 271:402–415
Acknowledgements
We are grateful to Dr. Virginia Aurora Herrera Valencia and Dr. Ignacio Islas Flores for critical reading and helpful comments on the manuscript. We also thank the handling editor and two reviewers for their constructive suggestions. This work was supported financially by the Centro de Investigación Científica de Yucatán (grant no. B06) and the Consejo Nacional de Ciencia y Tecnología (grant no. 24579) from México.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by A. Hirsch.
Nucleotide sequence data are in the GenBank database as accession nos. EF492518, EF492519, EF492520, EF492521, EF492522, EF492523, EF492524, EF492525, EF492526, EF492527, EF492528, EF492529, and EF492530 for Tg-4, Tg-6, Tg-9, Tg-10, Tg-12, Tg-13, Tg-67, Tg-1, Tg-2, Tg-20, Tg-34, Tg-36, and Tg-55, respectively.
Rights and permissions
About this article
Cite this article
Peraza-Echeverria, S., James-Kay, A., Canto-Canché, B. et al. Structural and phylogenetic analysis of Pto-type disease resistance gene candidates in banana. Mol Genet Genomics 278, 443–453 (2007). https://doi.org/10.1007/s00438-007-0262-9
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s00438-007-0262-9